Permutation test
A permutation test (also called re-randomization test or shuffle test) is an exact statistical hypothesis test making use of the proof by contradiction. A permutation test involves two or more samples. The null hypothesis is that all samples come from the same distribution . Under the null hypothesis, the distribution of the test statistic is obtained by calculating all possible values of the test statistic under possible rearrangements of the observed data. Permutation tests are, therefore, a form of resampling.
Permutation tests can be understood as surrogate data testing where the surrogate data under the null hypothesis are obtained through permutations of the original data.[1]
In other words, the method by which treatments are allocated to subjects in an experimental design is mirrored in the analysis of that design. If the labels are exchangeable under the null hypothesis, then the resulting tests yield exact significance levels; see also exchangeability. Confidence intervals can then be derived from the tests. The theory has evolved from the works of Ronald Fisher and E. J. G. Pitman in the 1930s.
Permutation tests should not be confused with randomized tests.[2]
Method
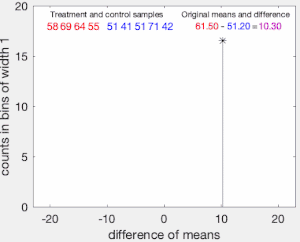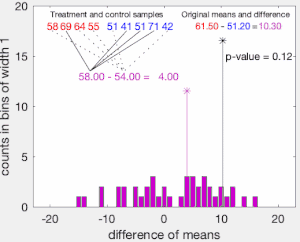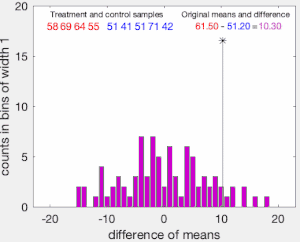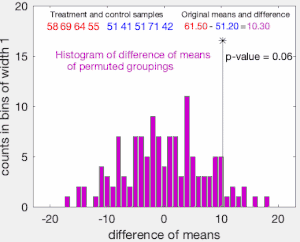
To illustrate the basic idea of a permutation test, suppose we collect random variables and for each individual from two groups and whose sample means are and , and that we want to know whether and come from the same distribution. Let and be the sample size collected from each group. The permutation test is designed to determine whether the observed difference between the sample means is large enough to reject, at some significance level, the null hypothesis H that the data drawn from is from the same distribution as the data drawn from .
The test proceeds as follows. First, the difference in means between the two samples is calculated: this is the observed value of the test statistic, .
Next, the observations of groups and are pooled, and the difference in sample means is calculated and recorded for every possible way of dividing the pooled values into two groups of size and (i.e., for every permutation of the group labels A and B). The set of these calculated differences is the exact distribution of possible differences (for this sample) under the null hypothesis that group labels are exchangeable (i.e., are randomly assigned).
The one-sided p-value of the test is calculated as the proportion of sampled permutations where the difference in means was greater than . The two-sided p-value of the test is calculated as the proportion of sampled permutations where the absolute difference was greater than . Many implementations of permutation tests require that the observed data itself be counted as one of the permutations so that the permutation p-value will never be zero.[3]
Alternatively, if the only purpose of the test is to reject or not reject the null hypothesis, one could sort the recorded differences, and then observe if is contained within the middle % of them, for some significance level . If it is not, we reject the hypothesis of identical probability curves at the significance level.
To exploit variance reduction with paired samples the paired permutation test needs to be applied, see paired difference test.
Relation to parametric tests
Permutation tests are a subset of non-parametric statistics. Assuming that our experimental data come from data measured from two treatment groups, the method simply generates the distribution of mean differences under the assumption that the two groups are not distinct in terms of the measured variable. From this, one then uses the observed statistic ( above) to see to what extent this statistic is special, i.e., the likelihood of observing the magnitude of such a value (or larger) if the treatment labels had simply been randomized after treatment.
In contrast to permutation tests, the distributions underlying many popular "classical" statistical tests, such as the t-test, F-test, z-test, and χ2 test, are obtained from theoretical probability distributions. Fisher's exact test is an example of a commonly used parametric test for evaluating the association between two dichotomous variables. When sample sizes are very large, the Pearson's chi-square test will give accurate results. For small samples, the chi-square reference distribution cannot be assumed to give a correct description of the probability distribution of the test statistic, and in this situation the use of Fisher's exact test becomes more appropriate.
Permutation tests exist in many situations where parametric tests do not (e.g., when deriving an optimal test when losses are proportional to the size of an error rather than its square). All simple and many relatively complex parametric tests have a corresponding permutation test version that is defined by using the same test statistic as the parametric test, but obtains the p-value from the sample-specific permutation distribution of that statistic, rather than from the theoretical distribution derived from the parametric assumption. For example, it is possible in this manner to construct a permutation t-test, a permutation test of association, a permutation version of Aly's test for comparing variances and so on.
The major drawbacks to permutation tests are that they
- Can be computationally intensive and may require "custom" code for difficult-to-calculate statistics. This must be rewritten for every case.
- Are primarily used to provide a p-value. The inversion of the test to get confidence regions/intervals requires even more computation.
Advantages
Permutation tests exist for any test statistic, regardless of whether or not its distribution is known. Thus one is always free to choose the statistic which best discriminates between hypothesis and alternative and which minimizes losses.
Permutation tests can be used for analyzing unbalanced designs[4] and for combining dependent tests on mixtures of categorical, ordinal, and metric data (Pesarin, 2001) [citation needed]. They can also be used to analyze qualitative data that has been quantitized (i.e., turned into numbers). Permutation tests may be ideal for analyzing quantitized data that do not satisfy statistical assumptions underlying traditional parametric tests (e.g., t-tests, ANOVA),[5] see PERMANOVA.
Before the 1980s, the burden of creating the reference distribution was overwhelming except for data sets with small sample sizes.
Since the 1980s, the confluence of relatively inexpensive fast computers and the development of new sophisticated path algorithms applicable in special situations made the application of permutation test methods practical for a wide range of problems. It also initiated the addition of exact-test options in the main statistical software packages and the appearance of specialized software for performing a wide range of uni- and multi-variable exact tests and computing test-based "exact" confidence intervals.
Limitations
An important assumption behind a permutation test is that the observations are exchangeable under the null hypothesis. An important consequence of this assumption is that tests of difference in location (like a permutation t-test) require equal variance under the normality assumption. In this respect, the classic permutation t-test shares the same weakness as the classical Student's t-test (the Behrens–Fisher problem). This can be addressed in the same way the classic t-test has been extended to handle unequal variances: by employing the Welch statistic with Satterthwaite adjustment to the degrees of freedom.[6] A third alternative in this situation is to use a bootstrap-based test. Statistician Phillip Good explains the difference between permutation tests and bootstrap tests the following way: "Permutations test hypotheses concerning distributions; bootstraps test hypotheses concerning parameters. As a result, the bootstrap entails less-stringent assumptions."[7] Bootstrap tests are not exact. In some cases, a permutation test based on a properly studentized statistic can be asymptotically exact even when the exchangeability assumption is violated.[8] Bootstrap-based tests can test with the null hypothesis and, therefore, are suited for performing equivalence testing.
Monte Carlo testing
An asymptotically equivalent permutation test can be created when there are too many possible orderings of the data to allow complete enumeration in a convenient manner. This is done by generating the reference distribution by Monte Carlo sampling, which takes a small (relative to the total number of permutations) random sample of the possible replicates. The realization that this could be applied to any permutation test on any dataset was an important breakthrough in the area of applied statistics. The earliest known references to this approach are Eden and Yates (1933) and Dwass (1957).[9][10] This type of permutation test is known under various names: approximate permutation test, Monte Carlo permutation tests or random permutation tests.[11]
After random permutations, it is possible to obtain a confidence interval for the p-value based on the Binomial distribution, see Binomial proportion confidence interval. For example, if after random permutations the p-value is estimated to be , then a 99% confidence interval for the true (the one that would result from trying all possible permutations) is .
On the other hand, the purpose of estimating the p-value is most often to decide whether , where is the threshold at which the null hypothesis will be rejected (typically ). In the example above, the confidence interval only tells us that there is roughly a 50% chance that the p-value is smaller than 0.05, i.e. it is completely unclear whether the null hypothesis should be rejected at a level .
If it is only important to know whether for a given , it is logical to continue simulating until the statement can be established to be true or false with a very low probability of error. Given a bound on the admissible probability of error (the probability of finding that when in fact or vice versa), the question of how many permutations to generate can be seen as the question of when to stop generating permutations, based on the outcomes of the simulations so far, in order to guarantee that the conclusion (which is either or ) is correct with probability at least as large as . ( will typically be chosen to be extremely small, e.g. 1/1000.) Stopping rules to achieve this have been developed[12] which can be incorporated with minimal additional computational cost. In fact, depending on the true underlying p-value it will often be found that the number of simulations required is remarkably small (e.g. as low as 5 and often not larger than 100) before a decision can be reached with virtual certainty.
Example tests
See also
Literature
Original references:
- Fisher, R.A. (1935) The Design of Experiments, New York: Hafner
- Pitman, E. J. G. (1937) "Significance tests which may be applied to samples from any population", Royal Statistical Society Supplement, 4: 119-130 and 225-32 (parts I and II). JSTOR 2984124 JSTOR 2983647
- Pitman, E. J. G. (1938). "Significance tests which may be applied to samples from any population. Part III. The analysis of variance test". Biometrika. 29 (3–4): 322–335. doi:10.1093/biomet/29.3-4.322.
Modern references:
- Collingridge, D.S. (2013). "A Primer on Quantitized Data Analysis and Permutation Testing". Journal of Mixed Methods Research. 7 (1): 79–95. doi:10.1177/1558689812454457. S2CID 124618343.
- Edgington, E. S., & Onghena, P. (2007) Randomization tests, 4th ed. New York: Chapman and Hall/CRC ISBN 9780367577711
- Good, Phillip I. (2005) Permutation, Parametric and Bootstrap Tests of Hypotheses, 3rd ed., Springer ISBN 0-387-98898-X
- Good, P (2002). "Extensions of the concept of exchangeability and their applications". Journal of Modern Applied Statistical Methods. 1 (2): 243–247. doi:10.22237/jmasm/1036110240.
- Lunneborg, Cliff. (1999) Data Analysis by Resampling, Duxbury Press. ISBN 0-534-22110-6.
- Pesarin, F. (2001). Multivariate Permutation Tests : With Applications in Biostatistics, John Wiley & Sons. ISBN 978-0471496700
- Welch, W. J. (1990). "Construction of permutation tests". Journal of the American Statistical Association. 85 (411): 693–698. doi:10.1080/01621459.1990.10474929.
Computational methods:
- Mehta, C. R.; Patel, N. R. (1983). "A network algorithm for performing Fisher's exact test in r x c contingency tables". Journal of the American Statistical Association. 78 (382): 427–434. doi:10.1080/01621459.1983.10477989.
- Mehta, C. R.; Patel, N. R.; Senchaudhuri, P. (1988). "Importance sampling for estimating exact probabilities in permutational inference". Journal of the American Statistical Association. 83 (404): 999–1005. doi:10.1080/01621459.1988.10478691.
- Gill, P. M. W. (2007). "Efficient calculation of p-values in linear-statistic permutation significance tests" (PDF). Journal of Statistical Computation and Simulation. 77 (1): 55–61. CiteSeerX 10.1.1.708.1957. doi:10.1080/10629360500108053. S2CID 1813706.
Current research on permutation tests
- Good, P.I. (2012) Practitioner's Guide to Resampling Methods.
- Good, P.I. (2005) Permutation, Parametric, and Bootstrap Tests of Hypotheses
- Hesterberg, T. C., D. S. Moore, S. Monaghan, A. Clipson, and R. Epstein (2005): Bootstrap Methods and Permutation Tests, software.
- Moore, D. S., G. McCabe, W. Duckworth, and S. Sclove (2003): Bootstrap Methods and Permutation Tests
- Simon, J. L. (1997): Resampling: The New Statistics.
- Yu, Chong Ho (2003): Resampling methods: concepts, applications, and justification. Practical Assessment, Research & Evaluation, 8(19). (statistical bootstrapping)
- Resampling: A Marriage of Computers and Statistics (ERIC Digests)
- Pesarin, F., Salmaso, L. (2010). Permutation Tests for Complex Data: Theory, Applications and Software. Wiley. https://books.google.com/books?id=9PWVTOanxPUC&hl=de
References
- ^ Moore, Jason H. "Bootstrapping, permutation testing and the method of surrogate data." Physics in Medicine & Biology 44.6 (1999): L11.
- ^ Onghena, Patrick (2017-10-30), Berger, Vance W. (ed.), "Randomization Tests or Permutation Tests? A Historical and Terminological Clarification", Randomization, Masking, and Allocation Concealment (1 ed.), Boca Raton, FL: Chapman and Hall/CRC, pp. 209–228, doi:10.1201/9781315305110-14, ISBN 978-1-315-30511-0, retrieved 2021-10-08
- ^ Phipson, Belinda; Smyth, Gordon K (2010). "Permutation p-values should never be zero: calculating exact p-values when permutations are randomly drawn". Statistical Applications in Genetics and Molecular Biology. 9 (1): Article 39. arXiv:1603.05766. doi:10.2202/1544-6115.1585. PMID 21044043. S2CID 10735784.
- ^ "Invited Articles" (PDF). Journal of Modern Applied Statistical Methods. 1 (2): 202–522. Fall 2011. Archived from the original (PDF) on May 5, 2003.
- ^ Collingridge, Dave S. (11 September 2012). "A Primer on Quantitized Data Analysis and Permutation Testing". Journal of Mixed Methods Research. 7 (1): 81–97. doi:10.1177/1558689812454457. S2CID 124618343.
- ^ Janssen, Arnold (1997). "Studentized Permutation Tests for Non-I.i.d. Hypotheses and the Generalized Behrens-Fisher Problem". Statistics & Probability Letters. 36 (1): 9–21. doi:10.1016/s0167-7152(97)00043-6.
- ^ Good, Phillip I. (2005). Resampling Methods: A Practical Guide to Data Analysis (3rd ed.). Birkhäuser. ISBN 978-0817643867.
- ^ Chung, EY; Romano, JP (2013). "Exact and asymptotically robust permutation tests". The Annals of Statistics. 41 (2): 487–507. arXiv:1304.5939. doi:10.1214/13-AOS1090.
- ^ Eden, T; Yates, F (1933). "On the validity of Fisher's z test when applied to an actual example of non-normal data. (With five text-figures.)". The Journal of Agricultural Science. 23 (1): 6–17. doi:10.1017/S0021859600052862. S2CID 84802682. Retrieved 3 June 2021.
- ^ Dwass, Meyer (1957). "Modified Randomization Tests for Nonparametric Hypotheses". Annals of Mathematical Statistics. 28 (1): 181–187. doi:10.1214/aoms/1177707045. JSTOR 2237031.
- ^ Thomas E. Nichols, Andrew P. Holmes (2001). "Nonparametric Permutation Tests For Functional Neuroimaging: A Primer with Examples" (PDF). Human Brain Mapping. 15 (1): 1–25. doi:10.1002/hbm.1058. hdl:2027.42/35194. PMC 6871862. PMID 11747097.
- ^ Gandy, Axel (2009). "Sequential implementation of Monte Carlo tests with uniformly bounded resampling risk". Journal of the American Statistical Association. 104 (488): 1504–1511. arXiv:math/0612488. doi:10.1198/jasa.2009.tm08368. S2CID 15935787.























![{\displaystyle \left[{\hat {p}}-z{\sqrt {\frac {0.05(1-0.05)}{10000}}},{\hat {p}}+z{\sqrt {\frac {0.05(1-0.05)}{10000}}}\right]=[0.045,0.055]}](https://wikimedia.org/api/rest_v1/media/math/render/svg/add2b4f210edaca5450abe5ec9b6199fb5af7d8f)






