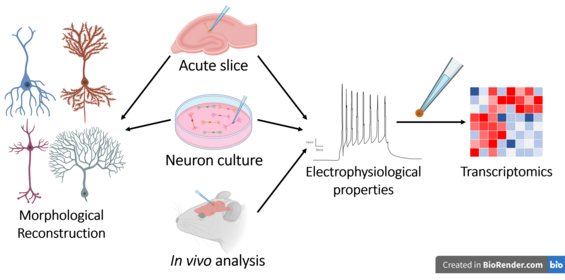
Patch-sequencing
Patch-sequencing (patch-seq) is a modification of patch-clamp technique that combines electrophysiological, transcriptomic and morphological characterization of individual neurons. In this approach, the neuron's cytoplasm is collected and processed for RNAseq after electrophysiological recordings are performed on it. The cell is simultaneously filled with a dye that allows for subsequent morphological reconstruction.
Background
Neuronal cell-typing requires simultaneous capturing of multiple data modalities
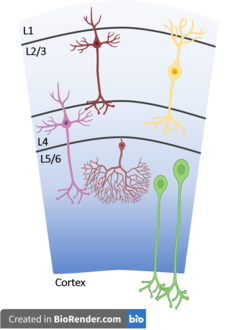
While a neuron's electrical properties are important when defining a cell type its morphology, types of neurotransmitters released, neurotransmitter receptors expressed at synapses, as well as the neuron's location in the nervous system and its local circuit are equally important. Neurons come in a huge diversity of shapes with many differences in cell bodies (soma), dendrites, and axons. The position of the dendrites determines which other neurons a cell receives its input from and their shape can have massive impacts on how a neurons responds to this input. Likewise the targets of a neuron's axon determine its outputs. The types of synapses formed between neurons' axons and dendrites are equally important as well.[1] For instance in the cortical microcircuit of the mammalian cortex, portrayed to the right, cells have highly specific projection patterns both within the local circuit as well as across cortical and non-cortical regions. Dendritic geometry influences the electrical behavior of neurons as well, having a massive influence on how dendrites process input in the form of postsynaptic potentials. Disordered geometry and projection patterns has been linked to a diverse set of psychiatric and neurological conditions including autism and schizophrenia though the behavioral relevance of these phenotypes is not yet understood.[2] Neuronal cell types appeared to often vary continuously between each other. Previous attempts at neuronal classification by morpho-electric properties have been limited by the use of incompatible methodologies and different cell line selection.[3]
With the advent of single-cell RNA-sequencing (scRNA-seq) it was hoped that there would exist genes that would be consistently expressed only in neurons with specific classically defined properties. These genes would serve as cell markers. This would provide a better means to delineate neuron types quickly and easily using only mRNA sequencing. However it appeared that scRNA-seq only served to reinforce the fact that overly rigid cell type definitions are not always the best way to characterize neurons.[4] Furthermore gene expression is dynamically regulated, varying over various time scales in response to activity in cell type specific ways to allow for neuronal plasticity.[5][6] Like other tissues, developmental processes also need to be considered.[7] Matching results from scRNA-seq to classically defined neuronal cell types is very challenging for all these reasons[8] and additionally single-cell RNA-seq has its own drawbacks for neuronal classification. While scRNA-seq enables the study of gene expression patterns from individual neurons, it disrupts the tissue for individual cell isolation and thus it is difficult to infer a neuron's original position in the tissue or observe its morphology. Linking the sequencing information to a neuronal subtype, defined previously by electrophysiological and morphological characteristics is a slow and complicated process.[9] The simultaneous capture and integration of multiple data types by patch-seq makes it ideal for neuronal classification, uncovering new correlations between gene expression, electrophysiological and morphological properties and neuronal function.[10] This makes patch-seq a truly interdisciplinary method, requiring collaboration between specialists in electrophysiology, sequencing, and imaging.[10]
Preparation and model system choice
Patch-seq can be done in any model system including cell culture for neurons. Neurons for culture may be collected from neuronal tissue then disassociated[11] or made from induced pluripotent stem cells (iPSC), neurons that have been grown out of human stem cell lines.[12] Cell culture preparation is the easiest to apply patch clamp to and give the experimenter control over what ligands the neuron is exposed to, for instance hormones or neurotransmitters. The benefit of total experimental control however also means the neurons are not subject to the natural environment they would be exposed to during development. As mention previously the position their dendrites and axons extend into as well as the neuron's position with a brain structure is incredibly important for understanding its role within a circuit. Many preparations exist for brain slices from different animal species.[13][14] Owing to the presence of cell or debris in the way of the pipette and a target cell the preparation will need to be slightly modified, often slight positive pressure is applied to the pipette to prevent any unintended seals from forming. If understanding how behavior is tied to the dynamics of the neuronal events is of interest it is possible to record in vivo as well. Though adapting patch clamp for in vivo studies can be very difficult for mechanical reasons especially during a behavioral task but has been done.[15] Automated in vivo patch clamp methods have been developed.[16] Very little difference exists between preparations for mammalian species though the greater diversity of neuronal sizes in non-human primate and primate cortex may necessitate using different tip diameters and pressures for forming seals without killing target neurons.[10] Patch-seq is also applied to non-neuronal studies such as pancreatic or cardiac cells.[17]
Patch-sequencing workflow
After choosing a model system and preparation type patch-seq experiments have a similar workflow. First a seal between the cell and the pipette is established so that recording and collection may take place. Cells can be filled with a fluorescent label for imaging during recording.[18] Following recording negative pressure is applied to capture the cytosol and often the nucleus for sequencing. This process is repeated until cells in the preparation have degraded and are not worth collecting data from. Post-hoc analysis of imaging data allows for morphological reconstruction. Like wise complex post-hoc processing of transcriptomic data is often required as well in order to handle a large number of confounds when collecting cytosolic contents from cells via the pipette.[19][20][21] In the initial stages, before forming the seal between the pipette and the cell, the tissue slices are prepared using a compresstome vibratome.To obtain thin sections of tissue, these devices are used. This device ensures that the target cells are accessible for patch clamping. The quality and precision of the tissue slicing are important for the success of the patch-seq experiment. The thickness and condition of these tissue slices influence the efficiency of cell targeting and the quality of the patch seal. An appropriate slice preparation is essential to the overall success of the patch-seq workflow.[22]
Forming the seal between pipette and cell
The patch pipette is designed for whole cell recording so its opening diameter is larger than experiments done to examine single ion-channels. For the most part standard patch clamp protocols may be used although there are some small situation dependent modifications to the pipette and the internal solution. An even wider diameter may be used to facilitate the aspiration of the inside of the cell into the pipette but it may need to be adjusted based on the target cell type.[23] Negative pressure is applied to enhance the seal which will be better for recording as well as prevent intra-cellular fluid leakage and contamination after or during collection of cell contents for sequencing. During recording biotin can be diffused into the cell via the pipette for imaging and later morphological reconstruction.[18]
Electrophysiological recording
Once a seal has been established cells are subject to different stimulation regimes using the voltage clamp, such as ramps, square pulses and noisy current injections. Features of the cell body's membrane are recorded including resting membrane potential and threshold potential. Features observed from generated action potentials (AP) such as AP width, AP amplitude, after-depolarization and after-hyperpolarization amplitude are also recorded.[3][9] Whole-cell recordings are performed using patch recoding pipettes filled with a small volume of intracellular solution (to avoid RNA dilution), calcium chelators, RNA carriers and RNase inhibitors.[10] The addition of RNase inhibitors, such as EGTA, enhances transcriptome analysis by preserving higher quality RNA from the samples.[9] Recording time can take between 1 and 15 minutes without affecting the neuron structure due to swelling, with lower values increasing throughput of the technique.[14] The data is recorded and analyzed with commercial or open-source software such as MIES,[3][24] PATCHMASTER,[25] pCLAMP,[26] WaveSurfer,[27] among others.
Neurons are pre-stimulated to verify their resting membrane potential and stabilize their baseline across and within experiments. Cells are then stimulated by ramp and square currents, their electrophysiological properties are recorded and measured. After stimulus the membrane potential must return to the baseline value for recordings to be consistent and robust. Negative pressure is used at the end of the recordings to return the membrane stability. Measurements need to satisfy these conditions to be considered for further analysis.[14] During recording cell viability needs to be maintained as being patch-clamped is stressful for the cell.
It is crucial to have healthy acute or live brain slices for electrophysiological recordings, as the health of the neuron significantly impacts the quality of the data obtained. Healthy brain slices are typically prepared using tools such as the Compresstome vibratome, ensuring optimal conditions for accurate and reliable recordings.[28]
Nucleus extraction
Negative pressure is used to move the nucleus near the pipette tip while moving the electrode near the center of the soma. The model system in question will affect the negative pressure to be applied. In human and non-human primates cell viability is more difficult to ensure. Larger variations in neuronal size compared to rodent models means greater variability in the amount of negative pressure needed to be applied to extract the cytosol and nucleus.[10] After retrieval the pipette is slowly retracted while maintaining negative pressure until the membrane surrounding the nucleus breaks off from the cell with the tip forming a membrane seal. The membrane seal traps the contents extracted in the pipette and prevents contamination during removal for sequencing before preparing the pipette for another recording. The construction of the seal can be observed electrically by the increase in resistance (MΩ).[14] The retrieval process is slow, often taking around ten minutes, as precision is needed to not burst the cell.[14]
Transcriptomics
RNA-seq analysis is performed using the nucleus and cytosol extracted from the recorded neurons.[9][14] RNA is amplified to full length cDNA and libraries are constructed for sequencing. Analyses including the nucleus not only have higher yields of mRNA but have increased data quality.[3]
Samples present a high degree of variability in the RNA content, in some cases including RNA contamination from adjacent cells such as astrocytes or other types of neurons. Quality is assessed by defined marker genes, indicating if the RNA content includes targets of the cell class of interest (on marker) and lacks any contamination markers (off marker).[14] Various metrics are used to judge the quality of collected RNA.
- Normalized marker sum (NMS) score: the ratio of on marker genes from the patch-seq cell relative to median expression of the same genes from an analogous data using cellular dissociation methods such as fluorescence-activated cell sorting (FACS). Cellular dissociation methods have reduced technical issues and acts as a standardization method.[19] The score measures the gene expression similarity pattern from the sample to a known cell class. Lower NMS scores indicate a reduced detection of on marker genes, more than an increase of off marker genes.[14]
- Contamination score: Indicates the likelihood of RNA contamination from near cells during extraction. Contamination may arise from pipette travel to the soma interacting with other neuronal processes.[19] It is calculated as the sum of the NMS score from all broad cell types that does not match the assigned cell class.
- Quality score: Measures the correlation between on and off marker genes from the patch-seq result with the average expression profile of cells analyzed using cellular dissociation methods of the same type.[14]
- Nucleus presence markers: Detection of nuclear specific genes (such as Malat1 in mammals)[19][29] and increased ratio of intronic reads provide evidence for nucleus incorporation in RNA-seq analysis.
Morphological reconstruction
Neuron shape and structure, such as dendritic/axonal arborization, axonal geometry, synaptic contacts and soma location, is used for neuronal type classification. In acute slice preparations or in vivo the laminar position is noted as it has important functional consequences as well. The staining by biotin is performed during neuron electrophysiological recording.[30] Alternatively rhodamine added outside the cell allows for imagine the living cell prior to recording.[31] Imaging is done by two-dimensional tiled images by bright field transmission and fluorescence channels on individual cells and then processed in commercial or open-source software.[14] Higher resolution images can be obtained from cultured neurons in comparison to acute slices, yielding higher quality morphological reconstructions.[10]
Post-hoc 3D reconstruction is made by image processing software such as TReMAP,[32] Mozak,[33] or Vaa3D.[34] Quality of the outcome depends on a variety of factors from the electrophysiological recording time to the nucleus extraction procedure. Reconstructed cells are then categorized in four levels of quality depending on their integrity and completeness of the structure. High quality if somas and processes are fully visible and proper digital reconstruction is possible, medium quality if somas and some processes are visible but are nor compatible with 3D reconstruction, insufficient axon where dendrites are filled with biotin but axons are weakly dyed, and failed fills that lack soma staining likely due to the subside of the structure post nuclear extraction.[14]
Downstream data analysis
Designing workflow for processing and combining the resulting multimodal data depends on the particular research question patch-seq is being applied to. In cell typing studies the data should be compared with existing scRNA-seq studies with larger sample sizes (in the order of thousands of cells compared to tens or hundreds) and therefore greater statistical power for cell type identification using transcriptomic data alone. Correlation based methods are sufficient for this step.[3][35] Dimensionality reduction methods such as T-distributed stochastic neighbor embedding or uniform manifold approximation and projection can then be used for visualization of the collected data's position on a reference atlas of higher quality scRNA-seq data.[36] Machine learning can be applied in order to relate the gene expression data to the morphological and electrophysiological data. Methods for doing so include autoencoders,[37] bottleneck networks,[38] or other rank reduction methods.[36] Including morphological data has proven to be challenging as it is a computer vision task, a notoriously complicated problem in machine learning. It is difficult to represent imaging data from the morphological reconstructions as a feature vector for including in the analysis.[10]
Applications
Patch-seq integrative dataset allows for a comprehensive characterization of cell types, particularly in neurons. Neuroscientists have applied this technique to a variety of experiments and protocols to discover new cell subtypes based on correlations between transcriptomic data and neuronal morpho-electric properties. Applying machine learning to patch-seq data it is then possible to study transcriptomic data and link it to their respective morpho-electric properties.[10] Having a confirmed ground truth for robust cell type classification allows researchers to look at the function of specific neuron types and subtypes in complex processes such as behavior, language and the underlying processes in neurological and psychiatric diseases. Comparison of proteomics with transcriptomics has shown that transcriptional data does not necessarily translate into the same protein expression[39] and likewise having the ability to look at the ground truth of a neuron's phenotype from classical classification methods combined with transcriptomic data is important for neuroscience. Patch-seq experiments have been found which support transcriptomic results but others have found cases where morphology of similar transcriptomically defined cell types in different brain regions did not match up.[10] The technique is particularly well suited for neuroscience but in general any tissue where it is of interest to simultaneous know electrophysiology, morphology, and transcriptomics would find use for patch-seq. For instance patch-seq has also been applied to non-neuronal tissues such as pancreatic islets for studying diabetes.[17]

- Targeted neuronal populations studies: Patch-seq excels compared to other methodologies in its capacity to measure and extract genetic information from targeted neurons without having to disrupt the sample. This allows for precise trace-back of the transcriptomic data to the neuron properties, such as its connectivity, the location in the slice, morphology. Correlation studies between these data types help identify differential expression genes across neuronal types and discover genetic markers for fast classification.[10]
- Compilation and integration of multimodal cell type databases: Patch-seq has been used to integrate neuron functional characteristics to the large transcriptional databases from single cell RNA-seq studies. Patch-seq can provide electrophysiological and morphological data, with morphology being less common due to the difficulty in achieving high quality standards.[14] Integration of the multimodal data is necessary as correct labeling of cell type is unobtainable from just one data type. Previous research has shown that transcriptomically similar neurons can have different morpho-electrical characteristics.[40]
- Molecular basis of morphological and functional diversity: Research into the molecular mechanisms that define the morphological and function diversity of neurons is ongoing. Patch-seq allows the identification of differential expressed genes between known distinct cell types. Small changes in gene expression can cause cells to display a different response to stimuli, underlining the importance of correlating this data for proper classification.[41]
Limitations
The most serious limitation to patch-seq is a limitation shared by patch clamp techniques generally, that being the requirement of high fidelity and dexterous manual labor. Patch clamping has been described as an art form and requires practice to perfect the technique. So not only is it labor-intensive but also takes years of training to prefect the technique.[42][43] To date the largest patch-seq study was done in acute cortical slices of mouse primary visual cortex. Just over 4000 neurons were patched and sequenced from and the effort required a huge amount of manual labor.[14] Most other data sets were collected from tens or hundreds of cells. Significantly less than cells collected for sequencing by dissociating the tissue.[10] Nevertheless no other electrophysiological recording's technique can match patch-seq's temporal resolution or ability to characterize specific ionic currents nor offer the capacity to produce a voltage clamp. For these reasons automating the technique is an area of active investigation by many laboratories around the world for applications using patch clamping including patch-seq.
Morphological reconstruction via digital imaging is another limitation of the technique with pass rates often lower than 50%.[14] The proper integration of the massive number of images with high quantities of noise, and the structural disruption caused by nucleus extraction, makes it a computational challenge in neuroscience. New automatic tracing methods are being developed, but quality remains low.[44]
Future directions
Integrating other sequencing technologies, proteomics, and single cell genetic manipulation
So far the only sequencing technology used has been mRNA sequencing. Including other modalities as well would increase the number of "omics" included in the data generated. For instance a technique for separating the mRNA from the DNA could allow for studying modifications on the DNA.[23] Chromatin accessibility could be judged from DNA methylation and using methylated DNA immunoprecipitation and histone acetylation and deacetylation from ChIP sequencing. This would enable study of epigenetics in patch-seq experiments. Likewise separating out proteins would be useful for judging protein abundances allowing for integration with proteomics. CRISPR could be included in the pipette and injected during recording to examine the effects of mutation at the single cell level.[10]
Auto-patching
The biggest bottle neck to the throughput of patch-seq is the labor required for patch clamping. Slowly automated patch clamp is beginning to become the norm but as of yet has not reached wide adoption. This is despite some auto-pathing rigs having a higher successfully attempted seal rate and data collection rate than humans. Automated patch clamping can either be done blind without imaging information instead relying on pressure sensors to provide input for algorithms guiding robotic rigs to form seals and record. Alternatively image guided algorithms exist as well allow for targeting of fluorescently labeled cells. Some systems are better suited to particular model systems and preparations.[43] [1]
See also
- Patch clamp
- Single-cell omics
- Single-cell sequencing
- RNA-Seq
- MAP-Seq
- Neuromorphology
- Neuronal tracing
- Electrophysiology
- Automated patch clamp
References
- ^ Larsen, Rylan S; Sjöström, P Jesper (2015-12-01). "Synapse-type-specific plasticity in local circuits". Current Opinion in Neurobiology. Circuit plasticity and memory. 35: 127–135. doi:10.1016/j.conb.2015.08.001. ISSN 0959-4388. PMC 5280068. PMID 26310110.
- ^ Pathania, M.; Davenport, E. C.; Muir, J.; Sheehan, D. F.; López-Doménech, G.; Kittler, J. T. (March 2014). "The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines". Translational Psychiatry. 4 (3): e374. doi:10.1038/tp.2014.16. ISSN 2158-3188. PMC 3966042. PMID 24667445.
- ^ a b c d e Gouwens, Nathan W.; Sorensen, Staci A.; Berg, Jim; Lee, Changkyu; Jarsky, Tim; Ting, Jonathan; Sunkin, Susan M.; Feng, David; Anastassiou, Costas; Barkan, Eliza; Bickley, Kris (2018-07-17). "Classification of electrophysiological and morphological types in mouse visual cortex". bioRxiv: 368456. doi:10.1101/368456. S2CID 91601091.
- ^ Cembrowski, Mark S.; Menon, Vilas (2018-06-01). "Continuous Variation within Cell Types of the Nervous System". Trends in Neurosciences. 41 (6): 337–348. doi:10.1016/j.tins.2018.02.010. ISSN 0166-2236. PMID 29576429. S2CID 4338758.
- ^ Hrvatin, Sinisa; Hochbaum, Daniel R.; Nagy, M. Aurel; Cicconet, Marcelo; Robertson, Keiramarie; Cheadle, Lucas; Zilionis, Rapolas; Ratner, Alex; Borges-Monroy, Rebeca; Klein, Allon M.; Sabatini, Bernardo L. (January 2018). "Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex". Nature Neuroscience. 21 (1): 120–129. doi:10.1038/s41593-017-0029-5. ISSN 1546-1726. PMC 5742025. PMID 29230054.
- ^ Yap, Ee-Lynn; Greenberg, Michael E. (2018-10-24). "Activity-regulated transcription: Bridging the gap between neural activity and behavior". Neuron. 100 (2): 330–348. doi:10.1016/j.neuron.2018.10.013. ISSN 0896-6273. PMC 6223657. PMID 30359600.
- ^ Fleming, Angeleen; Rubinsztein, David C. (2020-10-01). "Autophagy in Neuronal Development and Plasticity". Trends in Neurosciences. 43 (10): 767–779. doi:10.1016/j.tins.2020.07.003. ISSN 0166-2236. PMID 32800535. S2CID 221106496.
- ^ Johnson, Matthew B; Walsh, Christopher A (2017-02-01). "Cerebral cortical neuron diversity and development at single-cell resolution". Current Opinion in Neurobiology. Developmental neuroscience. 42: 9–16. doi:10.1016/j.conb.2016.11.001. ISSN 0959-4388. PMC 5316371. PMID 27888678.
- ^ a b c d Cadwell, Cathryn R; Palasantza, Athanasia; Jiang, Xiaolong; Berens, Philipp; Deng, Qiaolin; Yilmaz, Marlene; Reimer, Jacob; Shen, Shan; Bethge, Matthias; Tolias, Kimberley F; Sandberg, Rickard (February 2016). "Electrophysiological, transcriptomic and morphologic profiling of single neurons using Patch-seq". Nature Biotechnology. 34 (2): 199–203. doi:10.1038/nbt.3445. ISSN 1087-0156. PMC 4840019. PMID 26689543.
- ^ a b c d e f g h i j k l Lipovsek, Marcela; Bardy, Cedric; Cadwell, Cathryn R.; Hadley, Kristen; Kobak, Dmitry; Tripathy, Shreejoy J. (2021-02-03). "Patch-seq: Past, Present, and Future". Journal of Neuroscience. 41 (5): 937–946. doi:10.1523/JNEUROSCI.1653-20.2020. ISSN 0270-6474. PMC 7880286. PMID 33431632.
- ^ YAZAWA, Kazuto; KAIBARA, Muneshige; OHARA, Masahiro; KAMEYAMA, Masaki (1990). "An improved method for isolating cardiac myocytes useful for patch-clamp studies". The Japanese Journal of Physiology. 40 (1): 157–163. doi:10.2170/jjphysiol.40.157. ISSN 0021-521X. PMID 2163463.
- ^ Belinsky, Glenn S.; Rich, Matthew T.; Sirois, Carissa L.; Short, Shaina M.; Pedrosa, Erika; Lachman, Herbert M.; Antic, Srdjan D. (2014-01-01). "Patch-clamp recordings and calcium imaging followed by single-cell PCR reveal the developmental profile of 13 genes in iPSC-derived human neurons". Stem Cell Research. 12 (1): 101–118. doi:10.1016/j.scr.2013.09.014. ISSN 1873-5061. PMC 3947234. PMID 24157591.
- ^ Cadwell, Cathryn R.; Scala, Federico; Li, Shuang; Livrizzi, Giulia; Shen, Shan; Sandberg, Rickard; Jiang, Xiaolong; Tolias, Andreas S. (December 2017). "Multimodal profiling of single-cell morphology, electrophysiology, and gene expression using Patch-seq". Nature Protocols. 12 (12): 2531–2553. doi:10.1038/nprot.2017.120. ISSN 1750-2799. PMC 6422019. PMID 29189773.
- ^ a b c d e f g h i j k l m n Lee, Brian R.; Budzillo, Agata; Hadley, Kristen; Miller, Jeremy A.; Jarsky, Tim; Baker, Katherine; Hill, DiJon; Kim, Lisa; Mann, Rusty; Ng, Lindsay; Oldre, Aaron (2020-12-04). "Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization". bioRxiv: 2020.11.04.369082. doi:10.1101/2020.11.04.369082. S2CID 229297384.
- ^ Lee, Albert K.; Manns, Ian D.; Sakmann, Bert; Brecht, Michael (August 2006). "Whole-Cell Recordings in Freely Moving Rats". Neuron. 51 (4): 399–407. doi:10.1016/j.neuron.2006.07.004. PMID 16908406. S2CID 13524757.
- ^ Kodandaramaiah, Suhasa B.; Franzesi, Giovanni Talei; Chow, Brian Y.; Boyden, Edward S.; Forest, Craig R. (June 2012). "Automated whole-cell patch-clamp electrophysiology of neurons in vivo". Nature Methods. 9 (6): 585–587. doi:10.1038/nmeth.1993. ISSN 1548-7105. PMC 3427788. PMID 22561988.
- ^ a b Camunas-Soler, Joan; Dai, Xiao-Qing; Hang, Yan; Bautista, Austin; Lyon, James; Suzuki, Kunimasa; Kim, Seung K.; Quake, Stephen R.; MacDonald, Patrick E. (2020-05-05). "Patch-Seq Links Single-Cell Transcriptomes to Human Islet Dysfunction in Diabetes". Cell Metabolism. 31 (5): 1017–1031.e4. doi:10.1016/j.cmet.2020.04.005. ISSN 1550-4131. PMC 7398125. PMID 32302527.
- ^ a b Cadwell, Cathryn R; Scala, Federico; Li, Shuang; Livrizzi, Giulia; Shen, Shan; Sandberg, Rickard; Jiang, Xiaolong; Tolias, Andreas S (December 2017). "Multimodal profiling of single-cell morphology, electrophysiology, and gene expression using Patch-seq". Nature Protocols. 12 (12): 2531–2553. doi:10.1038/nprot.2017.120. ISSN 1754-2189. PMC 6422019. PMID 29189773.
- ^ a b c d Tripathy, Shreejoy J.; Toker, Lilah; Bomkamp, Claire; Mancarci, B. Ogan; Belmadani, Manuel; Pavlidis, Paul (2018). "Assessing Transcriptome Quality in Patch-Seq Datasets". Frontiers in Molecular Neuroscience. 11: 363. doi:10.3389/fnmol.2018.00363. ISSN 1662-5099. PMC 6187980. PMID 30349457.
- ^ Lipovsek, M.; Bardy, C.; Cadwell, C. R.; Hadley, K.; Kobak, D.; Tripathy, S. J. (2021). "Patch-seq: Past, Present, and Future". The Journal of Neuroscience. 41 (5): 937–946. doi:10.1523/JNEUROSCI.1653-20.2020. PMC 7880286. PMID 33431632.
- ^ Tripathy, Shreejoy J.; Toker, Lilah; Bomkamp, Claire; Mancarci, B. Ogan; Belmadani, Manuel; Pavlidis, Paul (2018). "Assessing Transcriptome Quality in Patch-Seq Datasets". Frontiers in Molecular Neuroscience. 11: 363. doi:10.3389/fnmol.2018.00363. PMC 6187980. PMID 30349457.
- ^ Mahfooz, Kashif; Ellender, Tommas J. (2021). "Combining Whole-Cell Patch-Clamp Recordings with Single-Cell RNA Sequencing". Patch Clamp Electrophysiology. Methods in Molecular Biology. Vol. 2188. pp. 179–189. doi:10.1007/978-1-0716-0818-0_9. ISBN 978-1-0716-0817-3. PMID 33119852. S2CID 226203486.
- ^ a b Lipovsek, Marcela; Browne, Lorcan; Grubb, Matthew S. (December 2020). "Protocol for Patch-Seq of Small Interneurons". STAR Protocols. 1 (3): 100146. doi:10.1016/j.xpro.2020.100146. ISSN 2666-1667. PMC 7757288. PMID 33377040.
- ^ Seeman, Stephanie C; Campagnola, Luke; Davoudian, Pasha A; Hoggarth, Alex; Hage, Travis A; Bosma-Moody, Alice; Baker, Christopher A; Lee, Jung Hoon; Mihalas, Stefan; Teeter, Corinne; Ko, Andrew L (2018-09-26). Slutsky, Inna; Marder, Eve; Sjöström, Per Jesper (eds.). "Sparse recurrent excitatory connectivity in the microcircuit of the adult mouse and human cortex". eLife. 7: e37349. doi:10.7554/eLife.37349. ISSN 2050-084X. PMC 6158007. PMID 30256194.
- ^ "HEKA Elektronik - Download the latest software release, hardware drivers and manuals". www.heka.com. Retrieved 2021-02-24.
- ^ "pCLAMP 11 Software Suite". Molecular Devices. Retrieved 2021-02-24.
- ^ "WaveSurfer | Janelia Research Campus". www.janelia.org. Retrieved 2021-02-24.
- ^ Fontaine, R.; Hodne, K.; Weltzien, F. A. (2018). "Healthy Brain-pituitary Slices for Electrophysiological Investigations of Pituitary Cells in Teleost Fish". Journal of Visualized Experiments (138). doi:10.3791/57790. PMC 6126815. PMID 30176004.
- ^ Hutchinson, John N.; Ensminger, Alexander W.; Clemson, Christine M.; Lynch, Christopher R.; Lawrence, Jeanne B.; Chess, Andrew (2007-02-01). "A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains". BMC Genomics. 8 (1): 39. doi:10.1186/1471-2164-8-39. ISSN 1471-2164. PMC 1800850. PMID 17270048.
- ^ Feldmeyer, Dirk; Egger, Veronica; Lübke, Joachim; Sakmann, Bert (1999-11-15). "Reliable synaptic connections between pairs of excitatory layer 4 neurones within a single 'barrel' of developing rat somatosensory cortex". The Journal of Physiology. 521 (Pt 1): 169–190. doi:10.1111/j.1469-7793.1999.00169.x. ISSN 0022-3751. PMC 2269646. PMID 10562343.
- ^ van den Hurk, Mark; Erwin, Jennifer A.; Yeo, Gene W.; Gage, Fred H.; Bardy, Cedric (2018-08-10). "Patch-Seq Protocol to Analyze the Electrophysiology, Morphology and Transcriptome of Whole Single Neurons Derived From Human Pluripotent Stem Cells". Frontiers in Molecular Neuroscience. 11: 261. doi:10.3389/fnmol.2018.00261. ISSN 1662-5099. PMC 6096303. PMID 30147644.
- ^ Zhou, Zhi; Liu, Xiaoxiao; Long, Brian; Peng, Hanchuan (2016-01-01). "TReMAP: Automatic 3D Neuron Reconstruction Based on Tracing, Reverse Mapping and Assembling of 2D Projections". Neuroinformatics. 14 (1): 41–50. doi:10.1007/s12021-015-9278-1. ISSN 1559-0089. PMID 26306866. S2CID 9636781.
- ^ Roskams, Jane; Popović, Zoran (2016-11-02). "Power to the People: Addressing Big Data Challenges in Neuroscience by Creating a New Cadre of Citizen Neuroscientists". Neuron. 92 (3): 658–664. doi:10.1016/j.neuron.2016.10.045. ISSN 0896-6273. PMID 27810012. S2CID 23704393.
- ^ Peng, Hanchuan; Ruan, Zongcai; Long, Fuhui; Simpson, Julie H.; Myers, Eugene W. (April 2010). "V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets". Nature Biotechnology. 28 (4): 348–353. doi:10.1038/nbt.1612. ISSN 1087-0156. PMC 2857929. PMID 20231818.
- ^ Fuzik, Janos; Rehman, Sabah; Girach, Fatima; Miklosi, Andras G.; Korchynska, Solomiia; Arque, Gloria; Romanov, Roman A.; Hanics, János; Wagner, Ludwig; Meletis, Konstantinos; Yanagawa, Yuchio (2019-12-17). "Brain-wide genetic mapping identifies the indusium griseum as a prenatal target of pharmacologically unrelated psychostimulants". Proceedings of the National Academy of Sciences. 116 (51): 25958–25967. Bibcode:2019PNAS..11625958F. doi:10.1073/pnas.1904006116. ISSN 0027-8424. PMC 6929682. PMID 31796600.
- ^ a b Kobak, Dmitry; Bernaerts, Yves; Weis, Marissa A.; Scala, Federico; Tolias, Andreas; Berens, Philipp (2020-04-14). "Sparse reduced-rank regression for exploratory visualization of paired multivariate datasets". bioRxiv: 302208. doi:10.1101/302208. S2CID 90792329.
- ^ Gala, Rohan; Gouwens, Nathan; Yao, Zizhen; Budzillo, Agata; Penn, Osnat; Tasic, Bosiljka; Murphy, Gabe; Zeng, Hongkui; Sümbül, Uygar (2019-11-05). "A coupled autoencoder approach for multi-modal analysis of cell types". arXiv:1911.05663 [q-bio.NC].
- ^ Bernaerts, Yves; Berens, Philipp; Kobak, Dmitry (2020-06-18). "Sparse Bottleneck Networks for Exploratory Analysis and Visualization of Neural Patch-seq Data". arXiv:2006.10411 [cs.LG].
- ^ Specht, Harrison; Emmott, Edward; Petelski, Aleksandra A.; Huffman, R. Gray; Perlman, David H.; Serra, Marco; Kharchenko, Peter; Koller, Antonius; Slavov, Nikolai (2021-01-27). "Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2". Genome Biology. 22 (1): 50. doi:10.1186/s13059-021-02267-5. ISSN 1474-760X. PMC 7839219. PMID 33504367.
- ^ Scala, F.; Kobak, D.; Shan, S.; Bernaerts, Y.; Laturnus, S.; Cadwell, C. R.; Hartmanis, L.; Froudarakis, E.; Castro, J.; Tan, Z. H.; Papadopoulos, S. (2019-04-11). "Neocortical layer 4 in adult mouse differs in major cell types and circuit organization between primary sensory areas". bioRxiv: 507293. doi:10.1101/507293. S2CID 91269384.
- ^ Oláh, Viktor János; Lukacsovich, David; Winterer, Jochen; Arszovszki, Antónia; Lőrincz, Andrea; Nusser, Zoltan; Földy, Csaba; Szabadics, János (2020). "Functional specification of CCK+ interneurons by alternative isoforms of Kv4.3 auxiliary subunits". eLife. 9. doi:10.7554/eLife.58515. ISSN 2050-084X. PMC 7269670. PMID 32490811.
- ^ Penner, Reinhold (1995), "A Practical Guide to Patch Clamping", Single-Channel Recording, Boston, MA: Springer US, pp. 3–30, doi:10.1007/978-1-4419-1229-9_1, ISBN 978-1-4419-1230-5, retrieved 2021-02-25
- ^ a b Suk, Ho-Jun; Boyden, Edward S.; van Welie, Ingrid (October 2019). "Advances in the automation of whole-cell patch clamp technology". Journal of Neuroscience Methods. 326: 108357. doi:10.1016/j.jneumeth.2019.108357. ISSN 0165-0270. PMID 31336060. S2CID 198192157.
- ^ Liu, Siqi; Zhang, Donghao; Song, Yang; Peng, Hanchuan; Cai, Weidong (2017-11-27). "Automated 3D Neuron Tracing with Precise Branch Erasing and Confidence Controlled Back-Tracking". bioRxiv: 109892. doi:10.1101/109892. S2CID 195961934.
