Newick format
| Filename extensions | .tree |
|---|---|
| Internet media type | text/x-nh |
| Initial release | 24 June 1986 |
| Type of format | graph-theoretical trees |
| Open format? | Yes |
In mathematics and phylogenetics, Newick tree format (or Newick notation or New Hampshire tree format) is a way of representing graph-theoretical trees with edge lengths using parentheses and commas. It was adopted by James Archie, William H. E. Day, Joseph Felsenstein, Wayne Maddison, Christopher Meacham, F. James Rohlf, and David Swofford, at two meetings in 1986, the second of which was at Newick's restaurant[1] in Dover, New Hampshire, US. The adopted format is a generalization of the format developed by Meacham in 1984 for the first tree-drawing programs in Felsenstein's PHYLIP package.[2]
Examples
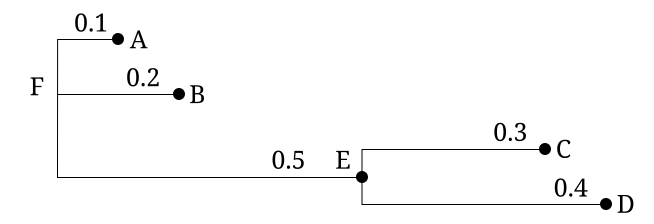
The following tree:
could be represented in Newick format in several ways
(,,(,)); no nodes are named (A,B,(C,D)); leaf nodes are named (A,B,(C,D)E)F; all nodes are named (:0.1,:0.2,(:0.3,:0.4):0.5); all but root node have a distance to parent (:0.1,:0.2,(:0.3,:0.4):0.5):0.0; all have a distance to parent (A:0.1,B:0.2,(C:0.3,D:0.4):0.5); distances and leaf names (popular) (A:0.1,B:0.2,(C:0.3,D:0.4)E:0.5)F; distances and all names ((B:0.2,(C:0.3,D:0.4)E:0.5)F:0.1)A; a tree rooted on a leaf node (rare)
Newick format is typically used for tools like PHYLIP and is a minimal definition for a phylogenetic tree.
Rooted, unrooted, and binary trees
When an unrooted tree is represented in Newick notation, an arbitrary node is chosen as its root. Whether rooted or unrooted, typically a tree's representation is rooted on an internal node and it is rare (but legal) to root a tree on a leaf node.
A rooted binary tree that is rooted on an internal node has exactly two immediate descendant nodes for each internal node. An unrooted binary tree that is rooted on an arbitrary internal node has exactly three immediate descendant nodes for the root node, and each other internal node has exactly two immediate descendant nodes. A binary tree rooted from a leaf has at most one immediate descendant node for the root node, and each internal node has exactly two immediate descendant nodes.
Grammar
A grammar for parsing the Newick format (roughly based on [3]):
The grammar nodes
Tree: The full input Newick Format for a single tree Subtree: an internal node (and its descendants) or a leaf node Leaf: a node with no descendants Internal: a node and its one or more descendants BranchSet: a set of one or more Branches Branch: a tree edge and its descendant subtree. Name: the name of a node Length: the length of a tree edge.
The grammar rules
Note, "|" separates alternatives.
Tree → Subtree ";"
Subtree → Leaf | Internal
Leaf → Name
Internal → "(" BranchSet ")" Name
BranchSet → Branch | Branch "," BranchSet
Branch → Subtree Length
Name → empty | string
Length → empty | ":" number
Whitespace (spaces, tabs, carriage returns, and linefeeds) within number is prohibited. Whitespace within string is often prohibited. Whitespace elsewhere is ignored. Sometimes the Name string must be of a specified fixed length; otherwise the punctuation characters from the grammar (semicolon, parentheses, comma, and colon) are prohibited. The Tree → Subtree ";" production is instead the Tree → Branch ";" production in those cases where having the entire tree descended from nowhere is permitted; this captures the replaced production as well because Length can be empty.
Note that when a tree having more than one leaf is rooted from one of its leaves, a representation that is rarely seen in practice, the root leaf is characterized as an Internal node by the above grammar. Generally, a root node labeled as Internal should be construed as actually internal if and only if it has at least two Branches in its BranchSet. One can make a grammar that formalizes this distinction by replacing the above Tree production rule with
Tree → RootLeaf ";" | RootInternal ";"
RootLeaf → Name | "(" Branch ")" Name
RootInternal → "(" Branch "," BranchSet ")" Name
The first RootLeaf production is for a tree with exactly one leaf. The second RootLeaf production is for rooting a tree from one of its two or more leaves.
Notes
- An unquoted string may not contain blanks, parentheses, square brackets, single_quotes, colons, semicolons, or commas. Underscore characters in unquoted strings are converted to blanks.[3]
- A string may also be quoted by enclosing it in single quotes. Single quotes in the original string are represented as two consecutive single quote characters.[3]
- Whitespace may appear anywhere except within an unquoted string or a Length
- Newlines may appear anywhere except within a string or a Length.
- Comments are enclosed in square brackets. They can appear anywhere newlines are allowed.[3] Comments starting with
&are generally computer-generated for additional data. Some dialects allow nested comments.
Dialects
New Hampshire X format
The New Hampshire X (NHX) format is an extension to Newick that adds key-value data (gene duplication, etc.) to Newick nodes. This is done by putting the additional data in brackets [&&NHX:key=value:...] in the node labels. The brackets are used because they represent comments in the Nexus file format, so any parser not understanding these additional information will ignore them.[4]
Extended Newick
While the standard Newick notation is limited to phylogenetic trees, Extended Newick (Perl Bio::PhyloNetwork) can be used to encode explicit phylogenetic networks.[5] In a phylogenetic network, which is a generalization of a phylogenetic tree, a node either represents a divergence event (cladogenesis) or a reticulation event such as hybridization, introgression, horizontal (lateral) gene transfer or recombination. Nodes that represent a reticulation event are duplicated, annotated by introducing the # symbol into the Newick format, and numbered consecutively (using integer values starting with 1).
For example, if leaf Y is the product of hybridisation (x) between lineages leading to C and D in the tree above,
|
|
one can express this situation by defining two trees in standard Newick notation
(A,B,((C,Y)c,D)e)f; and (A,B,(C,(Y,D)d)e)f; standard Newick, all nodes are named (internal nodes lowercase, leaves upper case)
or in extended Newick notation
(A,B,((C,(Y)x#H1)c,(x#H1,D)d)e)f; extended Newick, all nodes are named; 1 is the integer identifying the hybrid node x
The x#H1 here is a hybrid node. It will be joined by the program into a single node when drawn.
This is the picture drawn by Dendroscope for this example:

The production rules above is modified by the following for labelling hybrid nodes (in general, nodes representing reticulation events):[6]
Leaf → Name Hybrid Hybrid → empty | "#" Type integer -- The #i part is an obligatory identifier for a hybrid node Type → empty | string -- type of reticulation, e.g., H = hybridisation, LGT = lateral gene transfer, R = recombination.
In the visualization of LGT events, for a given reticulate node, one incoming edge is usually drawn as an "acceptor" edge and all other incoming edges are drawn as "transfer" edges. Some programs (e.g. Dendroscope and SplitsTree) allow exactly one copy of the reticulate node to be labeled with ## to indicate that it corresponds to the acceptor edge.
Extended Newick is backward-compatible: a hybrid node would simply be interpreted as a few strangely-named nodes for legacy parsers.
Rich Newick format
The Rich Newick format, also known as the Rice Newick format, is a further extension of Extended Newick.[7] It adds support for:
- Unrooted phylogenies. This is simply done by writing an unrooted tree as usual (i.e., pick an arbitrary root at a binary branch point) and prefixing
[&U]to the string.[&R], on the other hand, can be used to force a rooted tree. - Bootstrap values and probabilities. This is done by adding additional
:[bootstrap]:[prob]fields after the length; fields can be left empty as long as the colons are present. This may be backward-incompatible.
Ad hoc extensions
Some other programs, like NWX, uses comments starting with & to encode additional information in an ad hoc manner:[8]
- MrBayes and BEAST add additional information like probability, length in years, standard deviation for values to the nodes. They also use
[%U].
Visualization
Many tools have been published to visualize Newick tree data. Specific examples include the ETE toolkit ("Environment for Tree Exploration")[9] and T-REX.[10] Phylogenetic software packages such as SplitsTree and the tree-viewer Dendroscope as well as the online tree viewing tool IcyTree can handle standard and extended Newick notation, while the phylogenetic network software PhyloNet makes use of both the Extended Newick and Rich Newick format.
See also
- phyloXML
- T-REX (Webserver) allows handling phylogenetic trees and networks in the Newick format.
- Smart Game Format is an application of Newick format and is widely used for recording board games.
References
- ^ Newick's Lobster House home page
- ^ "The Newick tree format".
- ^ a b c d Olsen, Gary (30 August 1990). "Interpretation of "Newick's 8:45" Tree Format".
- ^ Zmasek, Christian M. (1999). "The New Hampshire X Format (NHX)" (PDF).
- ^ Cardona, Gabriel; Rosselló, Francesc; Valiente, Gabriel (27 March 2008). "A perl package and an alignment tool for phylogenetic networks". BMC Bioinformatics. 9: 175. doi:10.1186/1471-2105-9-175. ISSN 1471-2105. PMC 2330044. PMID 18371228.
- ^ Cardona, Gabriel; Rosselló, Francesc; Valiente, Gabriel (2008). "Extended Newick: it is time for a standard representation of phylogenetic networks". BMC Bioinformatics. 9: 532. doi:10.1186/1471-2105-9-532. PMC 2621367. PMID 19077301.
- ^ Barnett, Robert Matthew (16 February 2012). "Rich Newick Format". Rice University Wiki.
- ^ Yu, Guangchuang. "Chapter 1 Importing Tree with Data". Data Integration, Manipulation and Visualization of Phylogenetic Tree.
- ^ Huerta-Cepas, Jaime; Serra, François; Bork, Peer (June 2016). "ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data". Molecular Biology and Evolution. 33 (6): 1635–1638. doi:10.1093/molbev/msw046. ISSN 0737-4038. PMC 4868116. PMID 26921390.
- ^ Boc, Alix; Diallo, Alpha Boubacar; Makarenkov, Vladimir (July 2012). "T-REX: a web server for inferring, validating and visualizing phylogenetic trees and networks". Nucleic Acids Research. 40 (Web Server issue): W573–579. doi:10.1093/nar/gks485. ISSN 1362-4962. PMC 3394261. PMID 22675075.
External links
- Miyamoto and Goodman's Phylogram of Eutherian Mammals An example of a large phylogram with its Newick format representation.
- Phylogenetic tree (newick) viewer (By Huerta-Cepas et al. 2016)