Neurogenomics
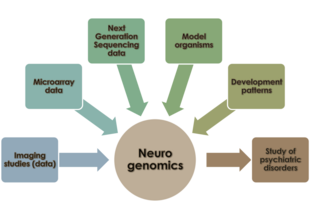
Neurogenomics is the study of how the genome of an organism influences the development and function of its nervous system.[1] This field intends to unite functional genomics and neurobiology in order to understand the nervous system as a whole from a genomic perspective.
The nervous system in vertebrates is made up of two major types of cells – neuroglial cells and neurons. Hundreds of different types of neurons exist in humans, with varying functions – some of them process external stimuli; others generate a response to stimuli; others organize in centralized structures (brain, spinal ganglia) that are responsible for cognition, perception, and regulation of motor functions. Neurons in these centralized locations tend to organize in giant networks and communicate extensively with each other. Prior to the availability of expression arrays and DNA sequencing methodologies, researchers sought to understand the cellular behaviour of neurons (including synapse formation and neuronal development and regionalization in the human nervous system) in terms of the underlying molecular biology and biochemistry, without any understanding of the influence of a neuron's genome on its development and behaviour. As our understanding of the genome has expanded, the role of networks of gene interactions in the maintenance of neuronal function and behaviour has garnered interest in the neuroscience research community. Neurogenomics allows scientists to study the nervous system of organisms in the context of these underlying regulatory and transcriptional networks. This approach is distinct from neurogenetics, which emphasizes the role of single genes without a network-interaction context when studying the nervous system.[2]
Approaches
Advent of high-throughput biology
In 1999, Cirelli & Tononi[3] first reported the association of genome-wide brain gene expression profiling (using microarrays) with a behavioural phenotype in mice. Since then, global brain gene expression data, derived from microarrays, has been aligned to various behavioural quantitative trait loci (QTLs) and reported in several publications.[4][5][6] However, microarray based approaches have their own problems that confound analysis – probe saturation can result in very small measurable variance of gene expression between genetically unique individuals,[7] and the presence of single nucleotide polymorphisms (SNPs) can result in hybridization artifacts.[8][9] Furthermore, due to their probe-based nature, microarrays can miss out on many types of transcripts (ncRNAs, miRNAs, and mRNA isoforms). Probes can also have species-specific binding affinities that can confound comparative analysis.
Notably, the association between behavioural patterns and high penetrance single gene loci falls under the purview of neurogenetics research, wherein the focus is to identify a simple causative relationship between a single, high penetrance gene and an observed function/behaviour. However, it has been shown that several neurological diseases tend to be polygenic, being influenced by multiple different genes and regulatory regions instead of one gene alone. There has hence been a shift from single gene approaches to network approaches for studying neurological development and diseases, a shift that has been greatly propelled by the advent of next generation sequencing methodologies.
Next-generation sequencing approaches
Twin studies have revealed that schizophrenia,[10] bipolar disorder,[11] autism spectrum disorder (ASD),[12][13] and attention deficit hyperactivity disorder[14] (ADHD) are highly heritable, genetically complex psychiatric disorders. However, linkage studies have largely failed at identifying causative variants for psychiatric disorders such as these, primarily because of their complex genetic architecture. Multiple low penetrance risk variants can be aggregated in affected individuals and families, and sets of causative variants could vary across families. Studies along these lines have determined a polygenic basis for several psychiatric disorders.[15] Several independently occurring de novo mutations in patients Alzheimer's disease have been found to disrupt a shared set of functional pathways involved with neuronal signalling, for example.[16] The quest to understand the causative biology of psychiatric disorders is hence greatly assisted by the ability to analyse entire genomes of affected and unaffected individuals in an unbiased manner.[17]
With the availability of massively parallel next generation sequencing methodologies, scientists have been able to look beyond the probe based captures of expressed genes. RNA-seq, for example, identifies 25-60% more expressed genes than microarrays do. In the upcoming field of neurogenomics, it is hoped that by understanding the genomic profiles of different parts of the brain, we might be able to improve our understanding of how the interactions between genes and pathways influence cellular function and development. This approach is expected to be able to identify the secondary gene networks that are disrupted in neurological disorders, subsequently assisting drug development stratagems for brain diseases.[18] The BRAIN initiative launched in 2013, for example, seeks to "inform the development of future treatments for brain disorders, including Alzheimer's disease, epilepsy, and traumatic brain injury" .
Rare variant association studies (RVAS) have highlighted the role of de novo mutations in several congenital and early-childhood-onset disorders like autism.[19][20] Several of these protein disrupting mutations have been able to be identified only with the aid of whole genome sequencing efforts, and validated with RNA-Seq. Additionally, these mutations are not statistically enriched in individual genes, but rather, exhibit patterns of statistical enrichment in groups of genes associated with networks regulating neurological development and maintenance. Such a discovery would have been impossible with prior gene-centric approaches (neurogenetics, behavioural neuroscience). Neurogenomics allows for a high-throughput system-based approach for understanding the polygenic basis of neuropsychiatric disorders.[17]
Imaging studies and optical mapping
When autism was identified as a distinct biological disorder in the 1980s, researchers found that autistic individuals showed a brain growth abnormality in the cerebellum in their early developmental years.[21] Subsequent research has indicated that 90% of autistic children have a larger brain volume than their peers by 2 to 4 years of age, and show an expansion in the white and gray matter content in the cerebrum.[22] The white and gray matter in the cerebrum is associated with learning and cognition respectively, and the formation of amyloid plaques in the white matter has been associated with Alzheimer's disease. These findings highlighted the influence of structural variance in the brain on psychiatric disorders, and have motivated the use of imaging technologies to map regions of divergence between healthy and diseased brains. Furthermore, while it may not always be possible to retrieve biological specimens from different areas live human brains, neuroimaging techniques offer a noninvasive means to understanding the biological basis of neurological disorders. It is hoped that an understanding of localization patterns of different psychiatric diseases could in turn inform network analysis studies in neurogenomics.
MRI
Structural Magnetic Resonance Imaging (MRI) can be used to identify the structural composition of the brain. Particularly in the context of neurogenomics, MRI has played an extensive role in the study of Alzheimer's disease over the past four decades. It was initially used to rule out other causes of dementia,[16] but recent studies indicated the presence of characteristic changes in patients with Alzheimer's disease. As a result, MRI scans are currently being used as a neuroimaging tool to help identify the temporal and spatial pathophysiology of Alzheimer's disease, such as specific cerebral alterations and amyloid imaging.[16]
The ease and non-invasive nature of MRI scans has motivated research projects that trace the development and onset of psychiatric diseases in the brain. Alzheimer disease has become a key candidate in this topographical approach to psychiatric diseases. For example, MRI scans are currently being used to track the resting and task-dependent functional profiles of brains in children with autosomal dominant Alzheimer disease.[23] These studies have found indications of early onset brain alterations in at-risk individuals for Alzheimer's disease.[16] The Autism Center of Excellence at University of California, San Diego, is also conducting MRI studies with children between 12 and 42 months, in the hopes of characterizing brain development abnormalities in children who present behavioural symptoms of autism.[24]
Additional research has indicated that there are specifics patterns of atrophy in the cerebrum (as a repercussion of neurodegeneration) in different neurological disorders and diseases. These disease-specific patterns of progression of atrophy can be identified with MRI scans, and provide a clinical phenotype context to neurogenomic research. The temporal information about disease progression provided by this approach can also potentially inform the interpretation of gene network-level perturbations in psychiatric diseases.[16]
Optical mapping
One prohibitive feature of 2nd generation sequencing methodologies is the upper limit on the genomic range accessible by mate-pairing. Optical mapping is an emerging methodology used to span large-scale variants that cannot usually be detected using paired end reads. This approach has been successfully applied to detect structural variants in oligodendroglioma, a type of brain cancer.[25] Recent work has also highlighted the versatility of optical maps in improving existing genome assemblies. Chromosomal rearrangements, microdeletions, and large-scale translocations have been associated with impaired neurological and cognitive function, for example in hereditary neuropathy and neurofibromatosis. Optical mapping can significantly improve variant detection and inform gene interaction network models for the diseased state in neurological disorders.
Studying other brain diseases
Apart from neurological disorders, there are additional diseases that manifest in the brain and have formed exemplar use-case scenarios for the application of brain imaging in network analysis. In a classic example of imaging-genomic analyses, a research study in 2012 compared MRI scans and gene expression profiles of 104 glioma patients in order to distinguish treatment outcomes and identify novel targetable genomic pathways in Glioblastoma Multiforme (GBM). Researchers found two distinct groups of patients with significantly different organization of white matter (invasive vs non-invasive). Subsequent pathway analysis of the gene expression data indicated mitochondrial dysfunction as the top canonical pathway in an aggressive, low-mortality GBM phenotype.[26]
Expansion of brain imaging approaches to other diseases can be used to rule out other medical illnesses while diagnosing psychiatric disorders, but cannot be used to inform the presence or absence of a psychiatric disorder.
Research developmental models
In humans
The current approaches in collecting gene expression data in human brains are to use either microarrays or RNA-seq. Currently, it is rare to gather "live" brain tissue – only when treatments involve brain surgery is there a chance that brain tissue is collected during the procedure. This is the case with epilepsy.
Currently, gene expression data is usually collected on post mortem brains and this is often a barrier to neurogenomics research in humans.[27][28] After death, the amount of time between death and when the data from the post mortem brain is collected is known as the post mortem interval (PMI). Since RNA degrades after death, a fresh brain is optimal – but not always available. This in turn can influence a variety of downstream analyses. Consideration should be taken of the following factors when working with 'omics data collected from post-mortem brains:
- Ideally, human brains should be controlled for PMIs for a given study.[29][30]
- The cause of death is also an important variable to consider in the collection of human brain samples for the purposes of neurogenomics research. For example, brain samples of individuals with clinical depression are often collected after suicide. Certain conditions of death, such as drug overdose or self-inflicted gunshot, will alter the expression of the brain.
- Another issue with studying gene expression in brains is the cellular heterogeneity of brain tissue samples. Bulk brain samples may vary in proportions of specific cell populations from case to case. This can impact the gene expression signatures and may significantly change differential expression analysis.
- One approach to address this issue is to use single cell RNA-seq. This would control for a specific cell type. However, this solution is only applicable where studies are not cell-type specific.[31]
Differential diagnosis also remains a critical pre-analytical confounder of cohort-wide studies of spectrum neurological disorders. Specifically, this has been noted to be a problem for Alzheimer's disease and autism spectrum disorder studies. Furthermore, as our understanding of the diverse symptoms and genomic underpinnings of various neurogenomic disorders improves, the diagnostic criteria itself undergoes rearrangements and review.[32]
Animal models
Ongoing genomics research in neurological disorders tends to use animal models (and corresponding gene homologs) to understand the network interactions underlying a particular disorder due to ethical issues surrounding the retrieval of biological specimens from live human brains. This, too, is not without its roadblocks.
Neurogenomic research with a model organism is contingent on the availability of a fully sequenced and annotated reference genome. Additionally, the RNA profiles (miRNA, ncRNA, mRNA) of the model organism need to be well catalogued, and any inferences applied from them to humans must have a basis in functional/sequence homology.[33]
Zebrafish
Zebrafish development relies on gene networks that are highly conserved among all vertebrates.[34] Additionally, with an extremely well annotated set of 12,000 genes and 1,000 early development mutants that are actually visible in the optically clear zebrafish embryos and larvae, zebrafish offer a sophisticated system for mutagenesis and real-time imaging of developing pathologies. This early development model has been employed to study the nervous system at cellular resolution.[35][36] The zebrafish model system has already been used to study neuroregeneration[37] and severe polygenic human diseases like cancer and heart disease.[38] Several zebrafish mutants with behavioural variations in response to cocaine and alcohol dosage have been isolated and can also form a basis for studying the pathogenesis of behavioural disorders.[39][40]
Rodent
Rodent models have been preeminent in studying human disorders. These models have been extensively annotated with gene homologs of several monogenic disorders in humans. Knockout studies of these homologs have led to expansion of our understanding of network interactions of genes in human tissues. For example, the FMR1 gene has been implicated with autism from a number of network studies.[41][42] Using a knockout of FMR1 in mice creates the model for Fragile X Syndrome, one of the disorders in the Autism spectrum.[43]
Mice xenografts are particularly useful for drug discovery,[44] and were extremely important in the discovery of early anti-psychotic drugs. The development of animal models for complex psychiatric diseases has also improved over the last few years. Rodent models have demonstrated behavioural phenotype changes resembling a positive schizophrenia state, either after genetic manipulation or after treatment with drugs that target the areas of the brain suspected to influence hyperactivity or neurodevelopment.[45] Interest has been generated in identifying the network disruptions mediated by these laboratory manipulations, and collection of genomic data from rodent studies has contributed significantly to a better understanding of the genomics of psychiatric diseases.
The first mouse brain transcriptome was generated in 2008.[46] Since then, extensive work has been done with building social-stress mice models to study the pathway level expression signatures of various psychiatric diseases. A recent paper simulated features of Post Traumatic Stress Disorder (PTSD) in mice, and profiled the entire transcriptome of these mice.[47] The authors found differential regulation in many biological pathways, some of which were implicated in anxiety disorders (hyperactivity, fear response), mood disorders, and impaired cognition. These findings are backed by extensive transcriptomic analyses of anxiety disorders, and expression level changes in biological pathways involved with fear learning and memory are thought to contribute to the behavioural manifestations of these disorders.[47] It is thought that functional enrichment of genes involved in long term synaptic potentiation, depression, and plasticity has an important role to play in the acquisition, consolidation, and maintenance of traumatic memories underlying anxiety disorders.[47][48]
Experimental mice models for psychiatric disorders
A common approach to using a mouse model is to apply an experimental treatment to a pregnant mouse in order to affect a whole litter. However, a key issue in the field is the treatment of litters in a statistical analysis. Most studies consider the total number of offspring produced as that may lead to an increase in statistical power. However, the correct way is to count by the number of litters and to normalize based on litter size. It was found that several autism studies incorrectly performed their statistical analyses based on total number of offspring instead of number of litters.[49]
Several anxiety disorders such as post-traumatic stress disorder (PTSD) involve heterogeneous changes in several different brain regions, such as the hippocampus, amygdala, and nucleus accumbens. The cellular encoding of traumatic events and the behavioral responses triggered by such events has been shown to lie primarily in changes in signaling molecules associated with synaptic transmission.
Global gene expression profiling of the various gene regions implicated in fear and anxiety processing, using mice models, has led to the identification of temporally and spatially distinct sets of differentially expressed genes. Pathway analysis of these genes has indicated possible roles in neurogenesis and anxiety-related behavioural responses, alongside other functional and phenotypic observations.[47]
Mice models for brain research have contributed significantly to drug development and increased our understanding of the genomic underpinnings of several neurological diseases in the last generation. Chlorpromazine, the first antipsychotic drug (discovered in 1951), was identified as a viable treatment option after it was shown to suppress response to aversive stimuli in rats in a behavioural screen.
Challenges
The modelling and assessment of latent symptoms (thoughts, verbal learning, social interactions, cognitive behaviour) remains a challenge when using model organisms to study psychiatric disorders with a complex genetic pathology. For example, a given genotype+phenotype in a mouse model must imitate the genomic underpinnings of a phenotype observed in a human.
This is a particularly crucial item of consideration in spectrum disorders such as autism. Autism is a disorder whose symptoms can be divided into two categories: (i) deficits of social interactions and (ii) repetitive behaviours and restricted interests. Since mice tend to be more social creatures amongst all members of the order Rodentia currently being used as model organisms, mice are generally used to model human psychiatric disorders as closely as possible. Particularly for autism, the following work-arounds are currently in place to emulate human behavioural symptoms:
- For the first diagnostic category of impaired social behaviour, mice are subject to a social assay intended to represent typical autistic social deficits. Normal social behaviour for mice includes sniffing, following, physical contact and allogrooming. Vocal communication could be used as well.
- There are a number of ways the second diagnostic category can be observed in mice. Examples of repetitive behaviours can include excessive circling, self-grooming and excessive digging. Usually these behaviours would be performed consistently within a long measurement of time (i.e. self-grooming for 10 minutes).[50]
- While repetitive behaviours are easily observable, it is difficult to characterize actual restricted interests of mice. One aspect of restricted interests of autistic individuals is the "insistence of sameness"—the concept that autistic individuals require their environment to remain consistent. If that environment should change, the individual would experience stress and anxiety. There has been reported success in confirming a mouse model of autism by changing the mouse's environment.[51]
In any of these experiments, the 'autistic' mice have a 'normal' socializing partner and the scientists observing the mice are unaware ("blind") to the genotypes of the mice.
Gene expression in the brain
The gene expression profile of the central nervous system (CNS) is unique. Eighty percent of all human genes are expressed in the brain; 5,000 of these genes are solely expressed in the CNS. The human brain has the highest amount of gene expression of all studied mammalian brains. In comparison, tissues outside of the brain will have more similar expression levels in comparison to their mammalian counterparts. One source of the increased expression levels in the human brain is from the non-protein coding region of the genome. Numerous studies have indicated that the human brain have a higher level of expression in regulatory regions in comparison to other mammalian brains. There is also notable enrichment for more alternative splicing events in the human brain.[2]
Spatial differences
Gene expression profiles also vary within specific regions of the brain. A microarray study showed that the transcriptome profile of the CNS clusters together based on region. A different study characterized the regulation of gene expression across 10 different regions based on their eQTL signals.[52] The cause of the varying expression profiles relates to function, neuron migration and cellular heterogeneity of the region. Even the three layers of the cerebral cortex have distinct expression profiles.[53]
A study completed at Harvard Medical School in 2014 was able to identify developmental lineages stemming from single base neuronal mutations. The researchers sequenced 36 neurons from the cerebral cortex of three normal individuals, and found that highly expressed genes, and neural associated genes, were significantly enriched for single-neuron SNVs. These SNVs, in turn, were found to be correlated with chromatin markers of transcription from fetal brain.[54]
Development patterns in humans
Gene expression of the brain changes throughout the different phases of life. The most significant levels of expression are found during early development, with the rate of gene expression being highest during fetal development. This results from the rapid growth of neurons in the embryo. Neurons at this stage are undergoing neuronal differentiation, cell proliferation, migration events and dendritic and synaptic development.[55] Gene expression patterns shift closer towards specialized functional profiles during embryonic development, however, certain developmental steps are still ongoing at parturition. Consequently, gene expression profiles of the two brain hemispheres appear asymmetrical at birth. At birth, gene expression profiles appear asymmetrical between brain hemispheres. As development continues, the gene expression profiles become similar between the hemispheres. Given a healthy adult, expression profiles stay relatively consistent from the late twenties into the late forties. From the fifties onwards, there is significant decrease in the expression of genes important for regular function. Despite this, there is an increase in the diversity of genes being expressed across the brain. This age related change in expression may be correlated with GC content. At later stages of life, there is an increase in the induction of low GC-content pivotal genes as well as an increase in the repression of high GC-content pivotal genes.[53] Another cause of the shift in gene diversity is the accumulation of mutations and DNA damage. Gene expression studies show that genes that accrue these age-related mutations are consistent between individuals in the aging population. Genes that are highly expressed at development decrease significantly at late stages in life, whereas genes that are highly repressed at development increase significantly at the late stages.[54]
Evolution of the mammalian brain
The evolution of Homo sapiens since the divergence from the primate common ancestor has shown a marked expansion in the size and complexity of the brain, especially in the cerebral cortex.[56][57][58][59] In comparison to primates, the human cerebral cortex has a larger surface area but differs only slightly in thickness. Many large scale studies in understanding the differences of the human brain from other species have indicated expansion of gene families and changes in alternative splicing to be responsible for the corollary increase in cognitive capabilities and cooperative behaviour in humans.[60][61] However, we are yet to determine the exact phenotypic consequences of all these changes. One difficulty is that only primates have developed subdivisions in their cerebral cortex, making the modeling of human specific neurological problems difficult to mimic in rodents.[58][62][63]
Sequence data is used to understand the evolutionary genetic changes which led to the development of the human CNS. We can then understand how the neurological phenotypes differ between species. Comparative genomics entails comparison of sequence data across a phylogeny to pinpoint the genotypic changes that occur within specific lineages, and understand how these changes might have arisen. The increase in high quality mammalian reference sequences generally makes comparative analysis better as it increases statistical power. However, the increase in number of species in a phylogeny does risk adding unnecessary noise as the alignments of the orthologous sequences usually decrease in quality. Furthermore, different classes of species will have significant differences in their phenotypes.[64]
Despite this, comparative genomics has allowed us to connect the genetic changes found in a phylogeny to specific pathways. In order to determine this, lineages are tested for the functional changes that accrue over time. This is often measured as a ratio of nonsynonymous substitutions to synonymous substitutions or the dN/dS ratio (sometimes, further abbreviated to ω). When the dN/dS ratio is greater than 1, this indicates positive selection. A dN/dS ratio equal to 1 is evidence of no selective pressures. A dN/dS ratio less than 1 indicates negative selection. For example, the conserved regions of the genome will generally have a dN/dS ratio of less than 1 since any changes to those positions will likely be detrimental.[65] Of the genes expressed in the human brain, it is estimated that 342 of them have a dN/dS ratio greater than 1 in the human lineage in comparison to other primate lineages.[64] This indicates positive selection on the human lineage for brain phenotypes. Understanding the significance of the positive selection is generally the next step. For example, ASPM, CDK5RAP2 and NIN are genes that are positively selected for on the human lineage and have been directly correlated with brain size. This finding may help elucidate why human brains are larger than other mammalian brains.[65]
Network level expression differences between species
It is thought that gene expression changes, being the ultimate response for any genetic changes, are a good proxy for understanding phenotypic differences within biological samples. Comparative studies have revealed a range of differences in the transcriptional controls between primates and rodents. For example, the gene CNTNAP2 is specifically enriched for in the prefrontal cortex. The mouse homolog of CNTNAP2 is not expressed in the mouse brain. CNTNAP2 has been implicated in cognitive functions of language as well as neurodevelopmental disorders such as Autism Spectrum Disorder. This suggests that the control of expression plays a significant role in the development in unique human cognitive function. As a consequence, a number of studies have investigated the brain specific enhancers. Transcription factors such as SOX5 have been found to be positively selected for on the human lineage. Gene expression studies in humans, chimpanzees and rhesus macaques, have identified human specific co-expression networks, and an elevation in gene expression in the human cortex in comparison to primates.[66]
Disorders
Neurogenomic disorders manifest themselves as neurological disorders with a complex genetic architecture and a non-Mendelian-like pattern of inheritance.[18] Some examples of these disorders include Bipolar disorder and Schizophrenia.[15] Several genes may be involved in the manifestation of the disorder, and mutations in such disorders are generally rare and de novo. Hence it becomes extremely unlikely to observe the same (potentially causative) variant in two unrelated individuals affected with the same neurogenomic disorder.[15] Ongoing research has implicated several de novo exonic variations and structural variations in Autism Spectrum Disorder (ASD), for example.[15] The allelic spectrum of the rare and common variants in neurogenomic disorders therefore necessitates a need for large cohort studies in order to effectively exclude low effect variants and identify the overarching pathways frequently mutated in the different disorders, rather than specific genes and specific high penetrance mutations.
Whole genome sequencing (WGS) and whole exome sequencing (WES) has been used in Genome Wide Association Studies (GWAS) to characterize genetic variants associated with neurogenomic disorders. However, the impact of these variants cannot always be verified because of the non-Mendelian inheritance patterns observed in several of these disorders.[15] Another prohibitive feature in network analysis is the lack of large-scale datasets for many psychiatric (neurogenomic) diseases. Since several diseases with neurogenomic underpinnings tend to have a polygenic basis, several nonspecific, rare, and partially penetrant de novo mutations in different patients can contribute to the same observed range of phenotypes, as is the case with Autism Spectrum Disorder and schizophrenia.[67] Extensive research in alcohol dependence has also highlighted the need for high-quality genomic profiling of large sample sets[68][69] when studying polygenic, spectrum disorders.
The 1000 Genomes Project was a successful demonstration of how a concerted effort to acquire representative genomic data from the broad spectrum of humans can result in identification of actionable biological insights for different diseases.[70] However, a large-scale initiative like this is still lacking in the field of neurogenomic disorders specifically.
Modelling psychiatric disorders in neurogenomics research – issues
One major GWAS study identified 13 new risk loci for schizophrenia.[71] Studying the impact of these candidates would ideally demonstrate a schizophrenia phenotype in animal models, which is usually difficult to observe due to its manifestation as a latent personality. This approach is able to determine the molecular impact the candidate gene. Ideally the candidate genes would have a neurological impact, which in turn would suggest that it plays a role in the neurological disorder. For example, in the aforementioned schizophrenia GWAS study, Ripke and colleagues[71] determined that these candidate genes were all involved in calcium signalling. Alternatively, one can study these variants in model organisms in the context of affected neurological function. It is important to note that the high penetrance variants of these disorders tend to be de novo mutations.
A further complication to studying neurogenomic disorders is the heterogeneous nature of the disorder. In many of these disorders, the mutations observed from case to case do not stay consistent. In autism, an affected individual may experience a large amount of deleterious mutations in gene X. A different affected individual may not have any significant mutations on gene X but have a large amount of mutations in gene Y. The alternative is to determine if gene X and gene Y impact the same biochemical pathway—one that influences a neurological function. A bioinformatics network analysis is one approach to this problem. Network analyses methodologies provide a generalized, systems overview of a molecular pathway.
One final complication to consider is the comorbidity of neurogenomic genes. Several disorders, especially at the more severe ends of the spectrum tend to be comorbid with each other. For example, more severe cases of ASD tend to be associated with intellectual disability (ID). This raises the question of whether or not there are true, unique ASD genes and unique ID genes or if there are just genes just associated with neurological function that can be mutated into an abnormal phenotype. One confounding factor may be the actual diagnostic category and methods of the spectrum disorders as symptoms between severe disorders may be similar. One study investigated the comorbid symptoms between groups of ID and ASD, and found no significant difference between the symptoms of ID children, ASD children with ID and ASD children without ID. Future research may help establish a more stringent genetic basis for the diagnoses of these disorders.
Network analysis

The main goal of network analysis in neurogenomics is to identify statistically significant nonrandom associations between genes that contain risk variants.[15] While several algorithm implementations of this approach already exist,[72][73] the general steps for network analysis remain the same.
- The analytical process starts out with the identification of a biological network based on experimental validation. This can be a gene co-expression network, or a protein-protein interaction (PPI) network. The nodes of the network will be clustered.
- Subsequently, a specific list of genes with known associations to a particular phenotype of interest is generated. This list could be determined by experimental data, agnostic of genetic studies in psychiatric disorders.[15] This is referred to as a 'hit list'.
- Genes that belong to the hit list as well as the biological network selected in the first step are marked as such.
- This is followed by a guilt-by-association (GBA) step. This means that clusters within the biological network that have a significant amount of genes from the hit list are investigated further using functional enrichment tools and database querying for the pathways in which these high scoring cluster genes participate[74]
- Thus the biological associations of the high-scoring, experimentally implicated cluster members are investigated, expanding the search area from beyond the initial hit list to include gene members of additional pathways that may have significant association with the initial biological network under consideration. This results in a set of candidate genes.[15]
The underlying principle of this approach is that the genes that cluster together, will also jointly affect the same molecular pathway. Again, they would ideally be part of a neurological function. The candidate genes can then be used to prioritize variants for wet lab validation.
Neuropharmacology
Historically, due to the behavioural stimulation manifested as a symptom in several the neurogenomic disorders, the therapies would rely mostly on anti-psychotics or antidepressants. These classes of medications would suppress common symptoms of the disorders, but with questionable efficacy. The biggest barrier to neruopharmacogenomic research was the cohort sizes. Given newly available large-cohort sequencing data, there has been a recent push to expand therapeutic options. The heterogenous nature of neurological diseases is the key motivation for personalized medicine approaches to their therapies. It is rare to find single high penetrance causative genes in neurological diseases. The genomic profiles understandably vary between cases, and logically, the therapies would need to vary between cases. Further complicating the issue is that many of these disorders are spectrum disorders. Their genetic etiology will vary within this spectrum. For example, severe ASD is associated with high penetrance de novo mutations. Milder forms of ASD is usually associated with a mixture of common variants.
The key issue then is the translation of these newly identified genetic variants (from Copy Number Variant studies, candidate gene sequencing and high throughput sequencing technologies) into an intervention for patients with neurogenomic disorders. One aspect will be if the neurological disorder are medically actionable (i.e. is there a simple metabolic pathway that a therapy can target). For example, specific cases of ASD have been associated with microdeletions on TMLHE gene. This gene codes for the enzyme of carnitine biosynthesis. Supplements to elevate carnitine levels appeared to alleviate certain ASD symptoms but the study was confounded by many influencing factors. As mentioned earlier, using a gene network approach will help identify relevant pathways of interest. Many neuropharmacogenomic approaches have focused on targeting the downstream products of these pathways.[75][76]
Blood brain barrier
Studies in animal models for several brain diseases has shown that the blood brain barrier (BBB) undergoes modification at many levels; for example, the surface glycoprotein composition can influence the types of HIV-1 strains transported by the BBB. The BBB has been found to be key in the onset of Alzheimer's disease.[77] It is extremely difficult, however, to be able to study this in humans due to obvious restrictions with accessing the brain and retrieving biological specimens for sequencing or morphological analysis. Mice models of the BBB and models of disease states have served well in conceptualizing the BBB as a regulatory interface between disease and good health in the brain.
Personalized neurobiology
The heterogenous nature of neurological diseases is the key motivation for personalized medicine approaches to their therapies.[75] Genomic samples of individual patients could be used to identify predictive factors, or to better understand the specific prognosis of a neurogenomic disease, and use this information to guide treatment options.[78] While there is a clear clinical utility to this approach, the adaptation of this approach is still nonexistent.
There are various issues prohibiting the application of personalized genomics to the assessment, diagnosis, and treatment of psychiatric disorders.
- Firstly, the causative network biology of several spectrum disorders with neurogenomic underpinnings is not fully understood yet, in spite of extensive studies conducted with disorders like Autism Spectrum[12][42] and schizophrenia.[10] Thus, the analytical validity of standing hypotheses concerning the etiology of neurogenomic disorders has still not been fully established and is subject to debate and controversy.
- The clinical validity of genetic variants that have shown to be highly correlated with specific neurogenomic disorders is often a major cause of concern.[78] The interpretation of these test results, and subsequent decision making, are a complicated undertaking given the polygenic nature of many of these disorders. Complicating things further, it has been shown that pre-emptive intervention in major psychiatric disorders does not always reduce the risk for the disorder.[79] Such intervention might not even be available for at-risk offspring of affected adults, thereby limiting the 'medical actionability' of the data.[78]
- Ethical concerns have also been raised regarding the safeguarding of personal genomic information, and how best to approach the burden of incidental findings and family risk assessment.
- Consanguinity and in-breeding can lead to selective enrichment of rare, otherwise low penetrance genetic mutations attributed to various symptoms of neurogenomic disorders. Thus, the interpretation of family-specific genetic mutations and/or network-level disruptions in the onset of a rare psychiatric disorder requires careful consideration of the motivations of participants included in the study.[78]
- That said, these issues can be addressed by effective education and counseling, and collection of genomic data from patients with psychiatric disorders should not be disqualified solely on this basis. The data itself serves as a dynamic health resource and can significantly further our understanding of the genomic basis of several psychiatric disorders.
See also
References
- ^ Boguski, Mark S.; Jones, Allan R. (2004-05-01). "Neurogenomics: at the intersection of neurobiology and genome sciences". Nature Neuroscience. 7 (5): 429–433. doi:10.1038/nn1232. ISSN 1097-6256. PMID 15114353. S2CID 14310435.
- ^ a b Jain, Kewal K. (2013-01-01). "Neurogenetics and Neurogenomics". Applications of Biotechnology in Neurology. Humana Press. pp. 7–16. doi:10.1007/978-1-62703-272-8_2. ISBN 9781627032711.
- ^ Cirelli, Chiara; Tononi, Giulio (1999). "Differences in gene expression during sleep and wakefulness". Annals of Medicine. 31 (2): 117–124. doi:10.3109/07853899908998787. PMID 10344584.
- ^ Matthews, Douglas B.; Bhave, Sanjiv V.; Belknap, John K.; Brittingham, Cynthia; Chesler, Elissa J.; Hitzemann, Robert J.; Hoffmann, Paula L.; Lu, Lu; McWeeney, Shannon (2005-09-01). "Complex genetics of interactions of alcohol and CNS function and behavior". Alcoholism: Clinical and Experimental Research. 29 (9): 1706–1719. doi:10.1097/01.alc.0000179209.44407.df. ISSN 0145-6008. PMID 16205371.
- ^ Hoffman, Paula L.; Miles, Michael; Edenberg, Howard J.; Sommer, Wolfgang; Tabakoff, Boris; Wehner, Jeanne M.; Lewohl, Joanne (2003-02-01). "Gene expression in brain: a window on ethanol dependence, neuroadaptation, and preference". Alcoholism: Clinical and Experimental Research. 27 (2): 155–168. doi:10.1097/01.ALC.0000060101.89334.11. ISSN 0145-6008. PMID 12605065.
- ^ Farris, Sean P.; Miles, Michael F. (2012-01-01). "Ethanol modulation of gene networks: implications for alcoholism". Neurobiology of Disease. 45 (1): 115–121. doi:10.1016/j.nbd.2011.04.013. ISSN 1095-953X. PMC 3158275. PMID 21536129.
- ^ Pozhitkov, Alex E.; Boube, Idrissa; Brouwer, Marius H.; Noble, Peter A. (2010-03-01). "Beyond Affymetrix arrays: expanding the set of known hybridization isotherms and observing pre-wash signal intensities". Nucleic Acids Research. 38 (5): e28. doi:10.1093/nar/gkp1122. ISSN 0305-1048. PMC 2836560. PMID 19969547.
- ^ Walter, Nicole A. R.; McWeeney, Shannon K.; Peters, Sandra T.; Belknap, John K.; Hitzemann, Robert; Buck, Kari J. (2007-09-01). "SNPs matter: impact on detection of differential expression". Nature Methods. 4 (9): 679–680. doi:10.1038/nmeth0907-679. ISSN 1548-7091. PMC 3410665. PMID 17762873.
- ^ Walter, Nicole A. R.; Bottomly, Daniel; Laderas, Ted; Mooney, Michael A.; Darakjian, Priscila; Searles, Robert P.; Harrington, Christina A.; McWeeney, Shannon K.; Hitzemann, Robert (2009-01-01). "High throughput sequencing in mice: a platform comparison identifies a preponderance of cryptic SNPs". BMC Genomics. 10: 379. doi:10.1186/1471-2164-10-379. ISSN 1471-2164. PMC 2743714. PMID 19686600.
- ^ a b Sullivan PF; Kendler KS; Neale MC (2003-12-01). "Schizophrenia as a complex trait: Evidence from a meta-analysis of twin studies". Archives of General Psychiatry. 60 (12): 1187–1192. doi:10.1001/archpsyc.60.12.1187. ISSN 0003-990X. PMID 14662550.
- ^ Smoller, Jordan W.; Finn, Christine T. (2003-11-15). "Family, twin, and adoption studies of bipolar disorder". American Journal of Medical Genetics Part C. 123C (1): 48–58. CiteSeerX 10.1.1.456.6790. doi:10.1002/ajmg.c.20013. ISSN 1552-4868. PMID 14601036. S2CID 29453951.
- ^ a b Rosenberg, Rebecca E.; Law, J. Kiely; Yenokyan, Gayane; McGready, John; Kaufmann, Walter E.; Law, Paul A. (2009-10-01). "Characteristics and concordance of autism spectrum disorders among 277 twin pairs". Archives of Pediatrics & Adolescent Medicine. 163 (10): 907–914. doi:10.1001/archpediatrics.2009.98. ISSN 1538-3628. PMID 19805709.
- ^ Frazier, Thomas W.; Thompson, Lee; Youngstrom, Eric A.; Law, Paul; Hardan, Antonio Y.; Eng, Charis; Morris, Nathan (2014-08-01). "A twin study of heritable and shared environmental contributions to autism". Journal of Autism and Developmental Disorders. 44 (8): 2013–2025. doi:10.1007/s10803-014-2081-2. ISSN 1573-3432. PMC 4104233. PMID 24604525.
- ^ Boomsma, Dorret; Busjahn, Andreas; Peltonen, Leena (2002-11-01). "Classical twin studies and beyond" (PDF). Nature Reviews Genetics. 3 (11): 872–882. doi:10.1038/nrg932. ISSN 1471-0056. PMID 12415317. S2CID 9318812.
- ^ a b c d e f g h Sullivan, Patrick F.; Daly, Mark J.; O'Donovan, Michael (2012-08-01). "Genetic architectures of psychiatric disorders: the emerging picture and its implications". Nature Reviews Genetics. 13 (8): 537–551. doi:10.1038/nrg3240. ISSN 1471-0056. PMC 4110909. PMID 22777127.
- ^ a b c d e Johnson, Keith A.; Fox, Nick C.; Sperling, Reisa A.; Klunk, William E. (2012-04-01). "Brain Imaging in Alzheimer Disease". Cold Spring Harbor Perspectives in Medicine. 2 (4): a006213. doi:10.1101/cshperspect.a006213. ISSN 2157-1422. PMC 3312396. PMID 22474610.
- ^ a b McCarroll, Steven A.; Feng, Guoping; Hyman, Steven E. (2014-06-01). "Genome-scale neurogenetics: methodology and meaning". Nature Neuroscience. 17 (6): 756–763. doi:10.1038/nn.3716. ISSN 1546-1726. PMC 4912829. PMID 24866041.
- ^ a b "Opinion: The Present and Future of Neurogenomics | The Scientist Magazine®". The Scientist. Retrieved 2016-02-23.
- ^ Malhotra, Dheeraj; Sebat, Jonathan (2012-03-16). "CNVs: harbingers of a rare variant revolution in psychiatric genetics". Cell. 148 (6): 1223–1241. doi:10.1016/j.cell.2012.02.039. ISSN 1097-4172. PMC 3351385. PMID 22424231.
- ^ McClellan, Jon; King, Mary-Claire (2010-06-23). "Genomic analysis of mental illness: a changing landscape". JAMA. 303 (24): 2523–2524. doi:10.1001/jama.2010.869. ISSN 1538-3598. PMID 20571020.
- ^ Courchesne, E.; Yeung-Courchesne, R.; Press, G. A.; Hesselink, J. R.; Jernigan, T. L. (1988-05-26). "Hypoplasia of cerebellar vermal lobules VI and VII in autism". The New England Journal of Medicine. 318 (21): 1349–1354. doi:10.1056/NEJM198805263182102. ISSN 0028-4793. PMID 3367935.
- ^ Courchesne, E.; Karns, C. M.; Davis, H. R.; Ziccardi, R.; Carper, R. A.; Tigue, Z. D.; Chisum, H. J.; Moses, P.; Pierce, K. (2001-07-24). "Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study". Neurology. 57 (2): 245–254. doi:10.1212/wnl.57.2.245. ISSN 0028-3878. PMID 11468308. S2CID 11234938.
- ^ Quiroz, Yakeel T.; Schultz, Aaron P.; Chen, Kewei; Protas, Hillary D.; Brickhouse, Michael; Fleisher, Adam S.; Langbaum, Jessica B.; Thiyyagura, Pradeep; Fagan, Anne M. (2015-08-01). "Brain Imaging and Blood Biomarker Abnormalities in Children With Autosomal Dominant Alzheimer Disease: A Cross-Sectional Study". JAMA Neurology. 72 (8): 912–919. doi:10.1001/jamaneurol.2015.1099. ISSN 2168-6157. PMC 4625544. PMID 26121081.
- ^ "UC San Diego Autism Center of Excellence". autism-center.ucsd.edu. Retrieved 2016-02-24.
- ^ Ray, Mohana; Goldstein, Steve; Zhou, Shiguo; Potamousis, Konstantinos; Sarkar, Deepayan; Newton, Michael A; Esterberg, Elizabeth; Kendziorski, Christina; Bogler, Oliver (2013-07-26). "Discovery of structural alterations in solid tumor oligodendroglioma by single molecule analysis". BMC Genomics. 14 (1): 505. doi:10.1186/1471-2164-14-505. PMC 3727977. PMID 23885787.
- ^ Colen, Rivka R.; Vangel, Mark; Wang, Jixin; Gutman, David A.; Hwang, Scott N.; Wintermark, Max; Jain, Rajan; Jilwan-Nicolas, Manal; Chen, James Y. (2014-01-01). "Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project". BMC Medical Genomics. 7: 30. doi:10.1186/1755-8794-7-30. ISSN 1755-8794. PMC 4057583. PMID 24889866.
- ^ Lipska, Barbara K.; Deep-Soboslay, Amy; Weickert, Cynthia Shannon; Hyde, Thomas M.; Martin, Catherine E.; Herman, Mary M.; Kleinman, Joel E. (2006-09-15). "Critical Factors in Gene Expression in Postmortem Human Brain: Focus on Studies in Schizophrenia". Biological Psychiatry. 60 (6): 650–658. doi:10.1016/j.biopsych.2006.06.019. PMID 16997002. S2CID 39379510.
- ^ Stan, Ana D.; Ghose, Subroto; Gao, Xue-Min; Roberts, Rosalinda C.; Lewis-Amezcua, Kelly; Hatanpaa, Kimmo J.; Tamminga, Carol A. (2006-12-06). "Human postmortem tissue: What quality markers matter?". Brain Research. 1123 (1): 1–11. doi:10.1016/j.brainres.2006.09.025. PMC 1995236. PMID 17045977.
- ^ Duric, Vanja; Banasr, Mounira; Stockmeier, Craig A.; Simen, Arthur A.; Newton, Samuel S.; Overholser, James C.; Jurjus, George J.; Dieter, Lesa; Duman, Ronald S. (2013-02-01). "Altered expression of synapse and glutamate related genes in post-mortem hippocampus of depressed subjects". International Journal of Neuropsychopharmacology. 16 (1): 69–82. doi:10.1017/S1461145712000016. ISSN 1461-1457. PMC 3414647. PMID 22339950.
- ^ Nagy, Corina; Maheu, Marissa; Lopez, Juan Pablo; Vaillancourt, Kathryn; Cruceanu, Cristiana; Gross, Jeffrey A.; Arnovitz, Mitchell; Mechawar, Naguib; Turecki, Gustavo (2015-05-01). "Effects of Postmortem Interval on Biomolecule Integrity in the Brain". Journal of Neuropathology & Experimental Neurology. 74 (5): 459–469. doi:10.1097/NEN.0000000000000190. ISSN 0022-3069. PMID 25868148.
- ^ Darmanis, Spyros; Sloan, Steven A.; Zhang, Ye; Enge, Martin; Caneda, Christine; Shuer, Lawrence M.; Gephart, Melanie G. Hayden; Barres, Ben A.; Quake, Stephen R. (2015-06-09). "A survey of human brain transcriptome diversity at the single cell level". Proceedings of the National Academy of Sciences. 112 (23): 7285–7290. Bibcode:2015PNAS..112.7285D. doi:10.1073/pnas.1507125112. ISSN 0027-8424. PMC 4466750. PMID 26060301.
- ^ Cicognola, Claudia; Chiasserini, Davide; Parnetti, Lucilla (2015-06-29). "Preanalytical Confounding Factors in the Analysis of Cerebrospinal Fluid Biomarkers for Alzheimer's Disease: The Issue of Diurnal Variation". Frontiers in Neurology. 6: 143. doi:10.3389/fneur.2015.00143. ISSN 1664-2295. PMC 4483516. PMID 26175714.
- ^ Rinkwitz, Silke; Mourrain, Philippe; Becker, Thomas S. (2011-02-01). "Zebrafish: an integrative system for neurogenomics and neurosciences". Progress in Neurobiology. 93 (2): 231–243. doi:10.1016/j.pneurobio.2010.11.003. ISSN 1873-5118. PMID 21130139. S2CID 34169168.
- ^ Cañestro, Cristian; Postlethwait, John H. (2007-05-15). "Development of a chordate anterior-posterior axis without classical retinoic acid signaling". Developmental Biology. 305 (2): 522–538. doi:10.1016/j.ydbio.2007.02.032. ISSN 0012-1606. PMID 17397819.
- ^ Tallafuss, Alexandra; Trepman, Alissa; Eisen, Judith S. (2009-12-01). "DeltaA mRNA and protein distribution in the zebrafish nervous system". Developmental Dynamics. 238 (12): 3226–3236. doi:10.1002/dvdy.22136. ISSN 1097-0177. PMC 2882441. PMID 19924821.
- ^ Russek-Blum, Niva; Gutnick, Amos; Nabel-Rosen, Helit; Blechman, Janna; Staudt, Nicole; Dorsky, Richard I.; Houart, Corinne; Levkowitz, Gil (2008-10-01). "Dopaminergic neuronal cluster size is determined during early forebrain patterning". Development. 135 (20): 3401–3413. doi:10.1242/dev.024232. ISSN 0950-1991. PMC 2692842. PMID 18799544.
- ^ Reimer, Michell M.; Sörensen, Inga; Kuscha, Veronika; Frank, Rebecca E.; Liu, Chong; Becker, Catherina G.; Becker, Thomas (2008-08-20). "Motor neuron regeneration in adult zebrafish". The Journal of Neuroscience. 28 (34): 8510–8516. doi:10.1523/JNEUROSCI.1189-08.2008. ISSN 1529-2401. PMC 6671064. PMID 18716209.
- ^ White, Richard; Rose, Kristin; Zon, Leonard (2013-09-01). "Zebrafish cancer: the state of the art and the path forward". Nature Reviews Cancer. 13 (9): 624–636. doi:10.1038/nrc3589. ISSN 1474-175X. PMC 6040891. PMID 23969693.
- ^ Darland, T.; Dowling, J. E. (2001). "Behavioral screening for cocaine sensitivity in mutagenized zebrafish". Proc. Natl. Acad. Sci. USA. 98 (20): 11691–11696. Bibcode:2001PNAS...9811691D. doi:10.1073/pnas.191380698. PMC 58791. PMID 11553778.
- ^ ; Lockwood, B., Bjerke, S., Kobayashi, K. & Guo, S. "Acute effects of alcohol on larval zebrafish: a genetic system for large-scale screening" Pharmacol. Biochem. Behav 2004; 77, 647–654
- ^ Bourgeron, Thomas (2015-09-01). "From the genetic architecture to synaptic plasticity in autism spectrum disorder". Nature Reviews Neuroscience. 16 (9): 551–563. doi:10.1038/nrn3992. ISSN 1471-003X. PMID 26289574. S2CID 12742356.
- ^ a b Just, Marcel Adam; Cherkassky, Vladimir L.; Keller, Timothy A.; Kana, Rajesh K.; Minshew, Nancy J. (2007-04-01). "Functional and Anatomical Cortical Underconnectivity in Autism: Evidence from an fMRI Study of an Executive Function Task and Corpus Callosum Morphometry". Cerebral Cortex. 17 (4): 951–961. doi:10.1093/cercor/bhl006. ISSN 1047-3211. PMC 4500121. PMID 16772313.
- ^ Oddi, D.; Crusio, W. E.; D'Amato, F. R.; Pietropaolo, S. (2013-08-15). "Monogenic mouse models of social dysfunction: Implications for autism". Behavioural Brain Research. SI:Neurobiology of Autism. 251: 75–84. doi:10.1016/j.bbr.2013.01.002. PMID 23327738. S2CID 10384899.
- ^ Gould, Stephen E.; Junttila, Melissa R.; de Sauvage, Frederic J. (2015-05-01). "Translational value of mouse models in oncology drug development". Nature Medicine. 21 (5): 431–439. doi:10.1038/nm.3853. ISSN 1078-8956. PMID 25951530. S2CID 5789096.
- ^ Jones, CA; Watson, DJG; Fone, KCF (2011-10-01). "Animal models of schizophrenia". British Journal of Pharmacology. 164 (4): 1162–1194. doi:10.1111/j.1476-5381.2011.01386.x. ISSN 0007-1188. PMC 3229756. PMID 21449915.
- ^ Mortazavi, Ali; Williams, Brian A.; McCue, Kenneth; Schaeffer, Lorian; Wold, Barbara (2008-07-01). "Mapping and quantifying mammalian transcriptomes by RNA-Seq". Nature Methods. 5 (7): 621–628. doi:10.1038/nmeth.1226. ISSN 1548-7105. PMID 18516045. S2CID 205418589.
- ^ a b c d Muhie, Seid; Gautam, Aarti; Meyerhoff, James; Chakraborty, Nabarun; Hammamieh, Rasha; Jett, Marti (2015-02-28). "Brain transcriptome profiles in mouse model simulating features of post-traumatic stress disorder". Molecular Brain. 8 (1): 14. doi:10.1186/s13041-015-0104-3. PMC 4359441. PMID 25888136.
- ^ Nutt, David J.; Malizia, Andrea L. (2004-01-01). "Structural and functional brain changes in posttraumatic stress disorder". The Journal of Clinical Psychiatry. 65 (Suppl 1): 11–17. ISSN 0160-6689. PMID 14728092.
- ^ Lazic, Stanley E; Essioux, Laurent (2013-03-22). "Improving basic and translational science by accounting for litter-to-litter variation in animal models". BMC Neuroscience. 14: 37. doi:10.1186/1471-2202-14-37. ISSN 1471-2202. PMC 3661356. PMID 23522086.
- ^ Crawley, Jacqueline N. (2012-09-01). "Translational animal models of autism and neurodevelopmental disorders". Dialogues in Clinical Neuroscience. 14 (3): 293–305. doi:10.31887/DCNS.2012.14.3/jcrawley. ISSN 1294-8322. PMC 3513683. PMID 23226954.
- ^ Gotham, Katherine; Bishop, Somer L.; Hus, Vanessa; Huerta, Marisela; Lund, Sabata; Buja, Andreas; Krieger, Abba; Lord, Catherine (2013-02-01). "Exploring the Relationship Between Anxiety and Insistence on Sameness in Autism Spectrum Disorders". Autism Research. 6 (1): 33–41. doi:10.1002/aur.1263. ISSN 1939-3806. PMC 4373663. PMID 23258569.
- ^ Ramasamy, Adaikalavan; Trabzuni, Daniah; Guelfi, Sebastian; Varghese, Vibin; Smith, Colin; Walker, Robert; De, Tisham; UK Brain Expression Consortium; North American Brain Expression Consortium (2014-10-01). "Genetic variability in the regulation of gene expression in ten regions of the human brain". Nature Neuroscience. 17 (10): 1418–1428. doi:10.1038/nn.3801. ISSN 1097-6256. PMC 4208299. PMID 25174004.
- ^ a b Naumova, Oksana Yu.; Lee, Maria; Rychkov, Sergei Yu.; Vlasova, Natalia V.; Grigorenko, Elena L. (2013-01-01). "Gene Expression in the Human Brain: The Current State of the Study of Specificity and Spatiotemporal Dynamics". Child Development. 84 (1): 76–88. doi:10.1111/cdev.12014. ISSN 1467-8624. PMC 3557706. PMID 23145569.
- ^ a b Lodato, Michael A.; Woodworth, Mollie B.; Lee, Semin; Evrony, Gilad D.; Mehta, Bhaven K.; Karger, Amir; Lee, Soohyun; Chittenden, Thomas W.; D'Gama, Alissa M. (2015-10-02). "Somatic mutation in single human neurons tracks developmental and transcriptional history". Science. 350 (6256): 94–98. Bibcode:2015Sci...350...94L. doi:10.1126/science.aab1785. ISSN 1095-9203. PMC 4664477. PMID 26430121.
- ^ Miller, Jeremy A.; Ding, Song-Lin; Sunkin, Susan M.; Smith, Kimberly A.; Ng, Lydia; Szafer, Aaron; Ebbert, Amanda; Riley, Zackery L.; Royall, Joshua J. (2014-04-10). "Transcriptional landscape of the prenatal human brain". Nature. 508 (7495): 199–206. Bibcode:2014Natur.508..199M. doi:10.1038/nature13185. ISSN 0028-0836. PMC 4105188. PMID 24695229.
- ^ Carroll, Sean B. (April 2003). "Genetics and the making of Homo sapiens". Nature. 422 (6934): 849–857. doi:10.1038/nature01495. PMID 12712196. S2CID 4333307.
- ^ Hill, Robert Sean; Walsh, Christopher A. (2005-09-01). "Molecular insights into human brain evolution". Nature. 437 (7055): 64–67. Bibcode:2005Natur.437...64H. doi:10.1038/nature04103. ISSN 1476-4687. PMID 16136130. S2CID 4406401.
- ^ a b Rakic, Pasko (2009-10-01). "Evolution of the neocortex: a perspective from developmental biology". Nature Reviews Neuroscience. 10 (10): 724–735. doi:10.1038/nrn2719. ISSN 1471-003X. PMC 2913577. PMID 19763105.
- ^ Geschwind, Daniel H.; Rakic, Pasko (2013-10-30). "Cortical evolution: judge the brain by its cover". Neuron. 80 (3): 633–647. doi:10.1016/j.neuron.2013.10.045. ISSN 1097-4199. PMC 3922239. PMID 24183016.
- ^ Calarco, John A.; Xing, Yi; Cáceres, Mario; Calarco, Joseph P.; Xiao, Xinshu; Pan, Qun; Lee, Christopher; Preuss, Todd M.; Blencowe, Benjamin J. (2007-11-15). "Global analysis of alternative splicing differences between humans and chimpanzees". Genes & Development. 21 (22): 2963–2975. doi:10.1101/gad.1606907. ISSN 0890-9369. PMC 2049197. PMID 17978102.
- ^ Zhang, Xiao-Ou; Yin, Qing-Fei; Wang, Hai-Bin; Zhang, Yang; Chen, Tian; Zheng, Ping; Lu, Xuhua; Chen, Ling-Ling; Yang, Li (2014-01-01). "Species-specific alternative splicing leads to unique expression of sno-lncRNAs". BMC Genomics. 15: 287. doi:10.1186/1471-2164-15-287. ISSN 1471-2164. PMC 4234469. PMID 24734784.
- ^ Somel, Mehmet; Liu, Xiling; Khaitovich, Philipp (2013-02-01). "Human brain evolution: transcripts, metabolites and their regulators". Nature Reviews Neuroscience. 14 (2): 112–127. doi:10.1038/nrn3372. ISSN 1471-003X. PMID 23324662. S2CID 12572416.
- ^ Qureshi, Irfan A.; Mehler, Mark F. (2012-08-01). "Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease". Nature Reviews Neuroscience. 13 (8): 528–541. doi:10.1038/nrn3234. ISSN 1471-003X. PMC 3478095. PMID 22814587.
- ^ a b Geschwind, Daniel H.; Rakic, Pasko (2013-10-30). "Cortical Evolution: Judge the Brain by Its Cover". Neuron. 80 (3): 633–647. doi:10.1016/j.neuron.2013.10.045. ISSN 0896-6273. PMC 3922239. PMID 24183016.
- ^ a b Enard, Wolfgang (2014-01-01). "Comparative genomics of brain size evolution". Frontiers in Human Neuroscience. 8: 345. doi:10.3389/fnhum.2014.00345. PMC 4033227. PMID 24904382.
- ^ Wang, Guang-Zhong; Konopka, Genevieve (2013-06-01). "Decoding human gene expression signatures in the brain". Transcription. 4 (3): 102–108. doi:10.4161/trns.24885. ISSN 2154-1272. PMC 4042582. PMID 23665540.
- ^ Kirov, G.; Pocklington, A. J.; Holmans, P.; Ivanov, D.; Ikeda, M.; Ruderfer, D.; Moran, J.; Chambert, K.; Toncheva, D. (2012-02-01). "De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia". Molecular Psychiatry. 17 (2): 142–153. doi:10.1038/mp.2011.154. ISSN 1476-5578. PMC 3603134. PMID 22083728.
- ^ Bierut, Laura J.; Agrawal, Arpana; Bucholz, Kathleen K.; Doheny, Kimberly F.; Laurie, Cathy; Pugh, Elizabeth; Fisher, Sherri; Fox, Louis; Howells, William (2010-03-16). "A genome-wide association study of alcohol dependence". Proceedings of the National Academy of Sciences. 107 (11): 5082–5087. Bibcode:2010PNAS..107.5082B. doi:10.1073/pnas.0911109107. ISSN 0027-8424. PMC 2841942. PMID 20202923.
- ^ Juraeva, Dilafruz; Treutlein, Jens; Scholz, Henrike; Frank, Josef; Degenhardt, Franziska; Cichon, Sven; Ridinger, Monika; Mattheisen, Manuel; Witt, Stephanie H. (2015-01-01). "XRCC5 as a risk gene for alcohol dependence: evidence from a genome-wide gene-set-based analysis and follow-up studies in Drosophila and humans". Neuropsychopharmacology. 40 (2): 361–371. doi:10.1038/npp.2014.178. ISSN 1740-634X. PMC 4443948. PMID 25035082.
- ^ The 1000 Genomes Project Consortium (2015-10-01). "A global reference for human genetic variation". Nature. 526 (7571): 68–74. Bibcode:2015Natur.526...68T. doi:10.1038/nature15393. ISSN 0028-0836. PMC 4750478. PMID 26432245.
{{cite journal}}: CS1 maint: numeric names: authors list (link) - ^ a b Ripke, Stephan; O'Dushlaine, Colm; Chambert, Kimberly; Moran, Jennifer L.; Kähler, Anna K.; Akterin, Susanne; Bergen, Sarah E.; Collins, Ann L.; Crowley, James J. (2013-10-01). "Genome-wide association analysis identifies 13 new risk loci for schizophrenia". Nature Genetics. 45 (10): 1150–1159. doi:10.1038/ng.2742. ISSN 1546-1718. PMC 3827979. PMID 23974872.
- ^ Lee, Phil H.; O'Dushlaine, Colm; Thomas, Brett; Purcell, Shaun M. (2012-07-01). "INRICH: interval-based enrichment analysis for genome-wide association studies". Bioinformatics. 28 (13): 1797–1799. doi:10.1093/bioinformatics/bts191. ISSN 1367-4811. PMC 3381960. PMID 22513993.
- ^ Morris, Andrew P; Voight, Benjamin F; Teslovich, Tanya M; Ferreira, Teresa; Segrè, Ayellet V; Steinthorsdottir, Valgerdur; Strawbridge, Rona J; Khan, Hassan; Grallert, Harald (2012-09-01). "Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes". Nature Genetics. 44 (9): 981–990. doi:10.1038/ng.2383. ISSN 1061-4036. PMC 3442244. PMID 22885922.
- ^ Gillis, Jesse; Pavlidis, Paul (2012). ""Guilt by Association" Is the Exception Rather Than the Rule in Gene Networks". PLOS Computational Biology. 8 (3): e1002444. Bibcode:2012PLSCB...8E2444G. doi:10.1371/journal.pcbi.1002444. PMC 3315453. PMID 22479173.
- ^ a b McMahon, Francis J.; Insel, Thomas R. (2012-06-07). "Pharmacogenomics and Personalized Medicine in Neuropsychiatry". Neuron. 74 (5): 773–776. doi:10.1016/j.neuron.2012.05.004. PMC 3407812. PMID 22681682.
- ^ Brandler, William M.; Sebat, Jonathan (2015-01-01). "From De Novo Mutations to Personalized Therapeutic Interventions in Autism". Annual Review of Medicine. 66 (1): 487–507. doi:10.1146/annurev-med-091113-024550. PMID 25587659.
- ^ Banks, William A. (2010-10-01). "Mouse models of neurological disorders: a view from the blood-brain barrier". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1802 (10): 881–888. doi:10.1016/j.bbadis.2009.10.011. ISSN 0006-3002. PMC 2891624. PMID 19879356.
- ^ a b c d Biesecker, Barbara Bowles; Peay, Holly Landrum (2013-08-01). "Genomic sequencing for psychiatric disorders: Promise and challenge". The International Journal of Neuropsychopharmacology. 16 (7): 1667–1672. doi:10.1017/S146114571300014X. ISSN 1461-1457. PMC 3703499. PMID 23575420.
- ^ Bunnik, Eline M; Schermer, Maartje HN; Janssens, A Cecile JW (2012-01-19). "The role of disease characteristics in the ethical debate on personal genome testing". BMC Medical Genomics. 5 (1): 4. doi:10.1186/1755-8794-5-4. PMC 3293088. PMID 22260407.