Lytic cycle
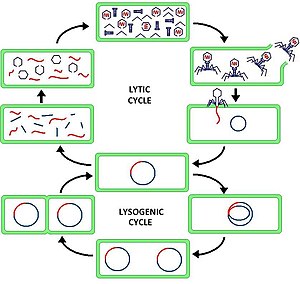
The lytic cycle (/ˈlɪtɪk/ LIT-ik) is one of the two cycles of viral reproduction (referring to bacterial viruses or bacteriophages), the other being the lysogenic cycle. The lytic cycle results in the destruction of the infected cell and its membrane. Bacteriophages that can only go through the lytic cycle are called virulent phages (in contrast to temperate phages).
In the lytic cycle, the viral DNA exists as a separate free floating molecule within the bacterial cell, and replicates separately from the host bacterial DNA, whereas in the lysogenic cycle, the viral DNA is integrated into the host genome. This is the key difference between the lytic and lysogenic cycles. However, in both cases the virus/phage replicates using the host DNA machinery.
Description
The lytic cycle is often separated into six stages: attachment, penetration, transcription, biosynthesis, maturation, and lysis.
- Attachment – the phage attaches itself to the surface of the host cell in order to inject its DNA into the cell
- Penetration – the phage injects its DNA into the host cell by penetrating through the cell membrane
- Transcription – the host cell's DNA is degraded and the cell's metabolism is directed to initiate phage biosynthesis
- Biosynthesis – the phage DNA replicates inside the cell, synthesizing new phage DNA and proteins
- Maturation – the replicated material assembles into fully formed viral phages (each made up of a head, a tail and tail fibers)
- Lysis - cell wall or membrane ruptures, disintegrating it and releasing the virus in the process
Attachment and penetration
To infect a host cell, the virus must first inject its own nucleic acid into the cell through the plasma membrane and (if present) the cell wall. The virus does so by either attaching to a receptor on the cell's surface or by simple mechanical force. The binding is due to electrostatic interactions and is influenced by pH and the presence of ions. The virus then releases its genetic material (either single- or double-stranded RNA or DNA) into the cell. In some viruses this genetic material is circular and mimics a bacterial plasmid. At this stage the cell becomes infected and can also be targeted by the immune system. It is mostly aided by receptors on the surface of the cell.[citation needed] The sequence of events that occur during initiation of bacteriophage infection, from adsorption (attachment) through DNA ejection from the virion into the host cell (penetration), was reviewed by Molineux.[1]
Transcription and biosynthesis
During the transcription and biosynthesis stages, the virus hijacks the cell's replication and translation mechanisms, using them to make more viruses. The virus's nucleic acid uses the host cell's metabolic machinery to make large amounts of viral components.[2]
In DNA viruses, the DNA transcribes itself into messenger RNA (mRNA) molecules that are then used to direct the cell's ribosomes. One of the first polypeptides to be translated destroys the host's DNA. In retroviruses (which inject an RNA strand), the enzyme reverse transcriptase transcribes the viral RNA into DNA, which is then transcribed again into RNA. Once the viral DNA has taken control it induces the host cell's machinery to synthesize viral DNA and proteins and begins to multiply.[citation needed]
The biosynthesis is (e.g. T4) regulated in three phases of mRNA production followed by a phase of protein production.[3]
- Early phase
- Enzymes modify the host's transcriptional process by RNA polymerase. Amongst other modifications, virus T4 changes the sigma factor of the host by producing an anti-sigma factor so that the host promotors are not recognized any more but now recognize T4 middle proteins. For protein synthesis Shine-Dalgarno subsequence GAGG dominates an early genes translation.[4]
- Middle phase
- Virus nucleic acid (DNA or RNA depending on virus type).[citation needed]
- Late phase
- Structural proteins including those for the head and the tail.[citation needed]
Maturation and lysis
About 25 minutes after initial infection, approximately 200 new virions (viral bodies) are formed. Once enough virions have matured and accumulated, specialized viral proteins are used to dissolve the cells' walls. The cell bursts (i.e. it undergoes lysis) due to high internal osmotic pressure that can no longer be constrained by the cell wall. This releases progeny virions into the surrounding environment, where they can go on to infect other cells and another lytic cycle begins.
Gene regulation biochemistry
There are three classes of genes in the phage genome that regulate whether the lytic or lysogenic cycles will emerge. The first class is the immediate early genes, the second is the delayed early genes and the third is the late genes. The following refers to the well-studied temperate phage lambda of E. coli.[citation needed]
- Immediate early genes: These genes are expressed from promoters recognized by the host RNA polymerase, and include Cro, cII, and N. CII is a transcription factor that stimulates expression of the main lysogenic repressor gene, cI, whereas Cro is a repressor for cI expression. The lysis-lysogeny decision is mainly influenced by the competition between Cro and CII, resulting in the determination of whether or not sufficient CI repressor is made. If so, CI represses the early promoters and the infection is shunted into the lysogenic pathway. N is an anti-termination factor that is needed for the transcription of the delayed early genes.
- Delayed early genes: These include the replication genes O and P and also Q, which encodes the anti-terminator responsible for transcription of all the late genes.
- Late genes:[more detail needed]
Q-mediated turn-on of late transcription begins about 6–8 min after infection if the lytic pathway is chosen. More than 25 genes are expressed from the single late promoter, resulting in four parallel biosynthetic pathways. Three of the pathways are for production of the three components of the virion: the DNA-filled head, the tail, and the side tail fibers. The virions self-assemble from these components, with the first virion appearing at about 20 min after infection. The fourth pathway is for lysis. In lambda 5 proteins are involved in lysis: the holin and antiholin from gene S, the endolysin from gene R and the spanin proteins from genes Rz and Rz1. In wild-type lambda, lysis occurs at about 50 min, releasing approximately 100 completed virions. The timing of lysis is determined by the holin and antiholin proteins, with the latter inhibiting the former. In overview, the holin protein accumulates in the cytoplasmic membrane until suddenly forming micron-scale holes, which triggers lysis. The endolysin R is released to the periplasm, where it attacks the peptidoglycan. The spanin proteins Rz and Rz1 accumulate in the cytoplasmic and outer membranes, respectively, and form a complex spanning the periplasm through the meshwork of the peptidoglycan. When the endolysin degrades the peptidoglycan, the spanin complexes are liberated and cause disruption of the outer membrane. Destruction of the peptidoglycan by the endolysin and disruption of the outer membrane by the spanin complex are both required for lysis in lambda infections.[citation needed]
Lysis inhibition: T4-like phages have two genes, rI and rIII, that inhibit the T4 holin, if the infected cell undergoes super-infection by another T4 (or closely related) virion. Repeated super-infection can cause the T4 infection to continue without lysis for hours, leading to accumulation of virions to levels 10-fold higher than normal.[5][better source needed]
References
- ^ Molineux, Ian J. (January 2006). "Fifty-three years since Hershey and Chase; much ado about pressure but which pressure is it?". Virology. 344 (1): 221–229. doi:10.1016/j.virol.2005.09.014.
- ^ "Ebola - Lytic or Lysogenic". Ebola-Cases.com. Retrieved 2023-01-26.
- ^ Madigan, Michael T.; Martinko, John M., eds. (2006). Brock biology of microorganisms (11 ed.). Prentice Hall. ISBN 978-0-13-144329-7.
- ^ Malys, N (2012). "Shine-Dalgarno sequence of bacteriophage T4: GAGG prevails in early genes". Molecular Biology Reports. 39 (1): 33–9. doi:10.1007/s11033-011-0707-4. PMID 21533668. S2CID 17854788.
- ^ "The Lytic Cycle of the T-Even Bacteriophage". nemetoadreviews.com. Retrieved January 9, 2018.

