HLA-A34
| HLA-A34 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||||||||
 HLA-A34 | ||||||||||||||||
| About | ||||||||||||||||
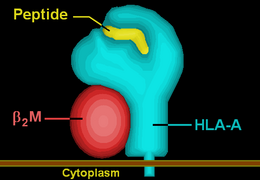
| Protein | transmembrane receptor/ligand | |||||||||||||||
| Structure | αβ heterodimer | |||||||||||||||
| Subunits | HLA-A*34--, β2-microglobulin | |||||||||||||||
| Older names | A10 | |||||||||||||||
| Subtypes | ||||||||||||||||
| ||||||||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||||||||
HLA-A34 (A34) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α34 subset of HLA-A α-chains. For A34, the alpha "A" chain are encoded by the HLA-A*34 allele group and the β-chain are encoded by B2M locus.[1] A34 and A*34 are almost synonymous in meaning. A34 is a split antigen of the broad antigen serotype A10. A34 is a sister serotype of A25, A26, A43, and A66.
A34 is most similar to A66.
Serotype
| A*34 | A34 | A10 | Sample |
| allele | % | % | size (N) |
| *3401 | 67 | 5 | 269 |
| *3402 | 89 | 2 | 996 |
A34 serotyping is modest.
A34 allele frequencies
A*3401
| freq | ||
| ref. | Population | (%) |
| [3] | Kimberly (Indig. Austr.) | 68.1 |
| [3] | Yuendumu (Indig. Austr.) | 44.0 |
| [3] | Groote Eylandt (Indig. Austr.) | 32.0 |
| [3] | Rabaul (New Britain, PNG) | 30.2 |
| [3] | West Schrader Ranges (PNG) | 29.2 |
| [3] | Cape York Penin. (Indig. Austr.) | 29.1 |
| [3] | Ami (Indig. Taiwan) | 21.9 |
| [3] | Goroka (E. Highlands, PNG) | 20.5 |
| [3] | New Caledonia | 15.5 |
| [3] | Madang (PNG) | 13.6 |
| [3] | Ivatan (N. Islands, Philippines) | 13.0 |
| [3] | Wanigela (PNG) | 12.7 |
| [3] | American Samoa | 10.0 |
| [3] | Wosera (PNG) | 9.0 |
| [3] | Riau Malay (Singapore) | 6.0 |
| [3] | Karimui Plateau (PNG) | 4.8 |
| [3] | Puyuma (Indig. Taiwan) | 4.0 |
| [3] | Javanese (Singapore) | 3.0 |
| [3] | Spain Catalonia Girona | 1.1 |
| [3] | Thai (Singapore) | 1.0 |
| [3] | Paiwan (Indig. Taiwan) | 1.0 |
| [3] | Taiwan Hakka | 0.9 |
| [3] | Kenya | 0.7 |
| [3] | Thailand | 0.7 |
| [3] | China South Han | 0.5 |
| [3] | India North Delhi | 0.5 |
| [3] | Chinese (Hong Kong) | 0.3 |
| [3] | Chinese (Singapore) | 0.3 |
| [3] | Zimbabwe Harare Shona | 0.2 |
A*3401 when found outside of Africa is primarily found in the South Asia, Austronesia and the South/Central part of the West Pacific Rim (WPR). It appears to have made it to Eastern Taiwan's indigenous tribes (Ami, Yami) but not more north of this region. It has not been detected in any sampling of Japan. Over most of the East Pacific Rim region where it is found it is limited to 1 or 2 common haplotypes in strong linkage disequilibrium. This indicates its presence in the WPR region is the result of recent migrations.[4]
The A*3401 migration from Africa is supported by its presence in East Africa and South Africa and by current models of human migration, this allele was likely represented in the first wave of immigrants. However, in areas were mixtures of these alleles are commonly found, Persian Gulf region and India, A*3401 is relatively uncommon, scarce, or absent. The exception is Saudi Arabia, in which A34 is at 2.8% and among Israeli Jews at 8.8%. Some reports that both A*3401 and A*3402 are in the region, but questionable whether these are perpetually maintained allele frequencies are simply recent migrants.
A*3402
| freq | ||
| ref. | Population | (%) |
| [3] | Natal Zulu (S. Africa) | 5.5 |
| [3] | Guinea Bissau | 5.4 |
| [3] | Bandiagara (Mali) | 3.6 |
| [3] | Shona (Zimbabwe) | 3.3 |
| [3] | Kenya | 2.8 |
| [3] | Tunisia Tunis | 2.8 |
| [3] | Uganda Kampala | 2.8 |
| [3] | Cameroon Bamileke | 2.6 |
| [3] | Senegal Niokholo Mandenka | 2.2 |
| [3] | Morocco Nador Metalsa | 2.1 |
| [3] | Zambia Lusaka | 1.2 |
| [3] | Sudanese | 1.0 |
| [3] | Israel Arab Druse | 0.5 |
| [3] | France South East | 0.4 |
| [3] | Jordan Amman | 0.3 |
| [3] | England Lancaster | 0.1 |
| [3] | Ireland Northern | 0.1 |
A*3402 is more frequently found in the west, it is found in Iberia and along the mediterranean, but its frequency is low, the exception may be in the Levant, but it is unclear whether this is A*3401 or A*3402.
A34-B Haplotypes
A34 is in strong linkage disequilibrium in many areas of the world, but particularly SE Asia and Oceania. The most prominent of these haplotypes is A34-Cw11(1)-B56. This haplotype is found from Western Australia to Taiwan to New Zealand indicating a recent genetic linkage between these peoples.
Another frequently found haplotype is the A34-B61 (A*3401:Cw*04:B*B4002) haplotype. This haplotype has a similar distribution as A34-B56.
These haplotypes indicate that long range/oversees migrations were taking place in Austronesias (late paleolithic) prehistory.
| freq | ||
| ref. | Population | (%) |
| [3] | Ami (Indig. Taiwan) | 19.9 |
| Highlanders (PNG) | 10.6 | |
| Maori (New Zealand) | 5.6 | |
| Indigenous Australia | 4.1 | |
| [3] | Puyuma (Indig. Taiwan) | 4.0 |
References
- ^ Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin". Tissue Antigens. 11 (2): 96–112. doi:10.1111/j.1399-0039.1978.tb01233.x. PMID 77067.
- ^ Allele Query Form IMGT/HLA - European Bioinformatics Institute
- ^ a b c d e f g h i j k l m n o p q r s t u v w x y z aa ab ac ad ae af ag ah ai aj ak al am an ao ap aq ar as at au av Middleton D, Menchaca L, Rood H, Komerofsky R (2003). "New allele frequency database". Tissue Antigens. 61 (5): 403–407. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
- ^ Probst, C.M. (2000). "HLA Polymorphism and Evaluation of European, African, and Amerindian Contribution to the White and Mulatto Populations". Human Biology. 72 (4): 597–617. JSTOR 41465861. PMID 11048789.
