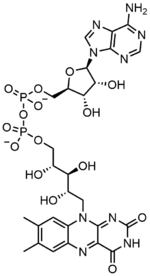
Flavin adenine dinucleotide
| |

| |
| Identifiers | |
|---|---|
3D model (JSmol) |
|
| 3DMet | |
| 1208946 | |
| ChEBI | |
| ChEMBL | |
| DrugBank | |
| ECHA InfoCard | 100.005.149 |
| EC Number |
|
| 108834 | |
| KEGG | |
| MeSH | Flavin-Adenine+Dinucleotide |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C27H33N9O15P2 | |
| Molar mass | 785.557 g·mol−1 |
| Appearance | White, vitreous crystals |
| log P | -1.336 |
| Acidity (pKa) | 1.128 |
| Basicity (pKb) | 12.8689 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
In biochemistry, flavin adenine dinucleotide (FAD) is a redox-active coenzyme associated with various proteins, which is involved with several enzymatic reactions in metabolism. A flavoprotein is a protein that contains a flavin group, which may be in the form of FAD or flavin mononucleotide (FMN). Many flavoproteins are known: components of the succinate dehydrogenase complex, α-ketoglutarate dehydrogenase, and a component of the pyruvate dehydrogenase complex.
FAD can exist in four redox states, which are the flavin-N(5)-oxide, quinone, semiquinone, and hydroquinone.[1] FAD is converted between these states by accepting or donating electrons. FAD, in its fully oxidized form, or quinone form, accepts two electrons and two protons to become FADH2 (hydroquinone form). The semiquinone (FADH·) can be formed by either reduction of FAD or oxidation of FADH2 by accepting or donating one electron and one proton, respectively. Some proteins, however, generate and maintain a superoxidized form of the flavin cofactor, the flavin-N(5)-oxide.[2][3]
History
Flavoproteins were first discovered in 1879 by separating components of cow's milk. They were initially called lactochrome due to their milky origin and yellow pigment.[4] It took 50 years for the scientific community to make any substantial progress in identifying the molecules responsible for the yellow pigment. The 1930s launched the field of coenzyme research with the publication of many flavin and nicotinamide derivative structures and their obligate roles in redox catalysis. German scientists Otto Warburg and Walter Christian discovered a yeast derived yellow protein required for cellular respiration in 1932. Their colleague Hugo Theorell separated this yellow enzyme into apoenzyme and yellow pigment, and showed that neither the enzyme nor the pigment was capable of oxidizing NADH on their own, but mixing them together would restore activity. Theorell confirmed the pigment to be riboflavin's phosphate ester, flavin mononucleotide (FMN) in 1937, which was the first direct evidence for enzyme cofactors.[5] Warburg and Christian then found FAD to be a cofactor of D-amino acid oxidase through similar experiments in 1938.[6] Warburg's work with linking nicotinamide to hydride transfers and the discovery of flavins paved the way for many scientists in the 40s and 50s to discover copious amounts of redox biochemistry and link them together in pathways such as the citric acid cycle and ATP synthesis.
Properties
Flavin adenine dinucleotide consists of two portions: the adenine nucleotide (adenosine monophosphate) and the flavin mononucleotide (FMN) bridged together through their phosphate groups. Adenine is bound to a cyclic ribose at the 1' carbon, while phosphate is bound to the ribose at the 5' carbon to form the adenine nucledotide. Riboflavin is formed by a carbon-nitrogen (C-N) bond between the isoalloxazine and the ribitol. The phosphate group is then bound to the terminal ribose carbon, forming a FMN. Because the bond between the isoalloxazine and the ribitol is not considered to be a glycosidic bond, the flavin mononucleotide is not truly a nucleotide.[7] This makes the dinucleotide name misleading; however, the flavin mononucleotide group is still very close to a nucleotide in its structure and chemical properties.

FAD can be reduced to FADH2 through the addition of 2 H+ and 2 e−. FADH2 can also be oxidized by the loss of 1 H+ and 1 e− to form FADH. The FAD form can be recreated through the further loss of 1 H+ and 1 e−. FAD formation can also occur through the reduction and dehydration of flavin-N(5)-oxide.[8] Based on the oxidation state, flavins take specific colors when in aqueous solution. flavin-N(5)-oxide (superoxidized) is yellow-orange, FAD (fully oxidized) is yellow, FADH (half reduced) is either blue or red based on the pH, and the fully reduced form is colorless.[9][10] Changing the form can have a large impact on other chemical properties. For example, FAD, the fully oxidized form is subject to nucleophilic attack, the fully reduced form, FADH2 has high polarizability, while the half reduced form is unstable in aqueous solution.[11] FAD is an aromatic ring system, whereas FADH2 is not.[12] This means that FADH2 is significantly higher in energy, without the stabilization through resonance that the aromatic structure provides. FADH2 is an energy-carrying molecule, because, once oxidized it regains aromaticity and releases the energy represented by this stabilization.
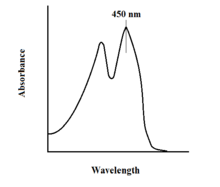
The spectroscopic properties of FAD and its variants allows for reaction monitoring by use of UV-VIS absorption and fluorescence spectroscopies. Each form of FAD has distinct absorbance spectra, making for easy observation of changes in oxidation state.[11] A major local absorbance maximum for FAD is observed at 450 nm, with an extinction coefficient of 11,300 M−1 cm−1.[13] Flavins in general have fluorescent activity when unbound (proteins bound to flavin nucleic acid derivatives are called flavoproteins). This property can be utilized when examining protein binding, observing loss of fluorescent activity when put into the bound state.[11] Oxidized flavins have high absorbances of about 450 nm, and fluoresce at about 515-520 nm.[9]
Chemical states
In biological systems, FAD acts as an acceptor of H+ and e− in its fully oxidized form, an acceptor or donor in the FADH form, and a donor in the reduced FADH2 form. The diagram below summarizes the potential changes that it can undergo.
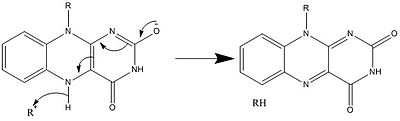
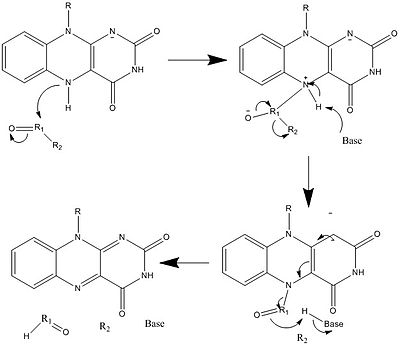
Along with what is seen above, other reactive forms of FAD can be formed and consumed. These reactions involve the transfer of electrons and the making/breaking of chemical bonds. Through reaction mechanisms, FAD is able to contribute to chemical activities within biological systems. The following pictures depict general forms of some of the actions that FAD can be involved in.
Mechanisms 1 and 2 represent hydride gain, in which the molecule gains what amounts to be one hydride ion. Mechanisms 3 and 4 radical formation and hydride loss. Radical species contain unpaired electron atoms and are very chemically active. Hydride loss is the inverse process of the hydride gain seen before. The final two mechanisms show nucleophilic addition and a reaction using a carbon radical.
Biosynthesis
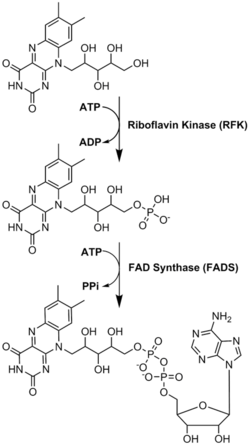
FAD plays a major role as an enzyme cofactor along with flavin mononucleotide, another molecule originating from riboflavin.[8] Bacteria, fungi and plants can produce riboflavin, but other eukaryotes, such as humans, have lost the ability to make it.[9] Therefore, humans must obtain riboflavin, also known as vitamin B2, from dietary sources.[14] Riboflavin is generally ingested in the small intestine and then transported to cells via carrier proteins.[9] Riboflavin kinase (EC 2.7.1.26) adds a phosphate group to riboflavin to produce flavin mononucleotide, and then FAD synthetase attaches an adenine nucleotide; both steps require ATP.[9] Bacteria generally have one bi-functional enzyme, but archaea and eukaryotes usually employ two distinct enzymes.[9] Current research indicates that distinct isoforms exist in the cytosol and mitochondria.[9] It seems that FAD is synthesized in both locations and potentially transported where needed.[11]
Function
Flavoproteins utilize the unique and versatile structure of flavin moieties to catalyze difficult redox reactions. Since flavins have multiple redox states they can participate in processes that involve the transfer of either one or two electrons, hydrogen atoms, or hydronium ions. The N5 and C4a of the fully oxidized flavin ring are also susceptible to nucleophilic attack.[15] This wide variety of ionization and modification of the flavin moiety can be attributed to the isoalloxazine ring system and the ability of flavoproteins to drastically perturb the kinetic parameters of flavins upon binding, including flavin adenine dinucleotide (FAD).
The number of flavin-dependent protein encoded genes in the genome (the flavoproteome) is species dependent and can range from 0.1% - 3.5%, with humans having 90 flavoprotein encoded genes.[16] FAD is the more complex and abundant form of flavin and is reported to bind to 75% of the total flavoproteome[16] and 84% of human encoded flavoproteins.[17] Cellular concentrations of free or non-covalently bound flavins in a variety of cultured mammalian cell lines were reported for FAD (2.2-17.0 amol/cell) and FMN (0.46-3.4 amol/cell).[18]
FAD has a more positive reduction potential than NAD+ and is a very strong oxidizing agent. The cell utilizes this in many energetically difficult oxidation reactions such as dehydrogenation of a C-C bond to an alkene. FAD-dependent proteins function in a large variety of metabolic pathways including electron transport, DNA repair, nucleotide biosynthesis, beta-oxidation of fatty acids, amino acid catabolism, as well as synthesis of other cofactors such as CoA, CoQ and heme groups. One well-known reaction is part of the citric acid cycle (also known as the TCA or Krebs cycle); succinate dehydrogenase (complex II in the electron transport chain) requires covalently bound FAD to catalyze the oxidation of succinate to fumarate by coupling it with the reduction of ubiquinone to ubiquinol.[11] The high-energy electrons from this oxidation are stored momentarily by reducing FAD to FADH2. FADH2 then reverts to FAD, sending its two high-energy electrons through the electron transport chain; the energy in FADH2 is enough to produce 1.5 equivalents of ATP[19] by oxidative phosphorylation. Some redox flavoproteins non-covalently bind to FAD like Acetyl-CoA-dehydrogenases which are involved in beta-oxidation of fatty acids and catabolism of amino acids like leucine (isovaleryl-CoA dehydrogenase), isoleucine, (short/branched-chain acyl-CoA dehydrogenase), valine (isobutyryl-CoA dehydrogenase), and lysine (glutaryl-CoA dehydrogenase).[20] Additional examples of FAD-dependent enzymes that regulate metabolism are glycerol-3-phosphate dehydrogenase (triglyceride synthesis) and xanthine oxidase involved in purine nucleotide catabolism.[21] Noncatalytic functions that FAD can play in flavoproteins include as structural roles, or involved in blue-sensitive light photoreceptors that regulate biological clocks and development, generation of light in bioluminescent bacteria.[20]
Flavoproteins
Flavoproteins have either an FMN or FAD molecule as a prosthetic group, this prosthetic group can be tightly bound or covalently linked. Only about 5-10% of flavoproteins have a covalently linked FAD, but these enzymes have stronger redox power.[11] In some instances, FAD can provide structural support for active sites or provide stabilization of intermediates during catalysis.[20] Based on the available structural data, the known FAD-binding sites can be divided into more than 200 types.[22]
90 flavoproteins are encoded in the human genome; about 84% require FAD, and around 16% require FMN, whereas 5 proteins require both to be present.[17] Flavoproteins are mainly located in the mitochondria because of their redox power.[17] Of all flavoproteins, 90% perform redox reactions and the other 10% are transferases, lyases, isomerases, ligases.[16]
Oxidation of carbon-heteroatom bonds
Carbon-nitrogen
Monoamine oxidase (MAO) is an extensively studied flavoenzyme due to its biological importance with the catabolism of norepinephrine, serotonin and dopamine. MAO oxidizes primary, secondary and tertiary amines, which nonenzymatically hydrolyze from the imine to aldehyde or ketone. Even though this class of enzyme has been extensively studied, its mechanism of action is still being debated. Two mechanisms have been proposed: a radical mechanism and a nucleophilic mechanism. The radical mechanism is less generally accepted because no spectral or electron paramagnetic resonance evidence exists for the presence of a radical intermediate. The nucleophilic mechanism is more favored because it is supported by site-directed mutagenesis studies which mutated two tyrosine residues that were expected to increase the nucleophilicity of the substrates.[23]
Carbon-oxygen
Glucose oxidase (GOX) catalyzes the oxidation of β-D-glucose to D-glucono-δ-lactone with the simultaneous reduction of enzyme-bound flavin. GOX exists as a homodimer, with each subunit binding one FAD molecule. Crystal structures show that FAD binds in a deep pocket of the enzyme near the dimer interface. Studies showed that upon replacement of FAD with 8-hydroxy-5-carba-5-deaza FAD, the stereochemistry of the reaction was determined by reacting with the re face of the flavin. During turnover, the neutral and anionic semiquinones are observed which indicates a radical mechanism.[23]
Carbon-sulfur
Prenylcysteine lyase (PCLase) catalyzes the cleavage of prenylcysteine (a protein modification) to form an isoprenoid aldehyde and the freed cysteine residue on the protein target. The FAD is non-covalently bound to PCLase. Not many mechanistic studies have been done looking at the reactions of the flavin, but the proposed mechanism is shown below. A hydride transfer from the C1 of the prenyl moiety to FAD is proposed, resulting in the reduction of the flavin to FADH2. COformED IS a carbocation that is stabilized by the neighboring sulfur atom. FADH2 then reacts with molecular oxygen to restore the oxidized enzyme.[23]
Carbon-carbon
UDP-N-acetylenolpyruvylglucosamine Reductase (MurB) is an enzyme that catalyzes the NADPH-dependent reduction of enolpyruvyl-UDP-N-acetylglucosamine (substrate) to the corresponding D-lactyl compound UDP-N-acetylmuramic acid (product). MurB is a monomer and contains one FAD molecule. Before the substrate can be converted to product, NADPH must first reduce FAD. Once NADP+ dissociates, the substrate can bind and the reduced flavin can reduce the product.[23]
Thiol/disulfide chemistry
Glutathione reductase (GR) catalyzes the reduction of glutathione disulfide (GSSG) to glutathione (GSH). GR requires FAD and NADPH to facilitate this reaction; first a hydride must be transferred from NADPH to FAD. The reduced flavin can then act as a nucleophile to attack the disulfide, this forms the C4a-cysteine adduct. Elimination of this adduct results in a flavin-thiolate charge-transfer complex.[23]
Electron transfer reactions
Cytochrome P450 type enzymes that catalyze monooxygenase (hydroxylation) reactions are dependent on the transfer of two electrons from FAD to the P450. Two types of P450 systems are found in eukaryotes. The P450 systems that are located in the endoplasmic reticulum are dependent on a cytochrome P-450 reductase (CPR) that contains both an FAD and an FMN. The two electrons on reduced FAD (FADH2) are transferred one at a time to FMN and then a single electron is passed from FMN to the heme of the P450.[24]
The P450 systems that are located in the mitochondria are dependent on two electron transfer proteins: An FAD containing adrenodoxin reductase (AR) and a small iron-sulfur group containing protein named adrenodoxin. FAD is embedded in the FAD-binding domain of AR.[25][26] The FAD of AR is reduced to FADH2 by transfer of two electrons from NADPH that binds in the NADP-binding domain of AR. The structure of this enzyme is highly conserved to maintain precisely the alignment of electron donor NADPH and acceptor FAD for efficient electron transfer.[26] The two electrons in reduced FAD are transferred one a time to adrenodoxin which in turn donates the single electron to the heme group of the mitochondrial P450.[27]
The structures of the reductase of the microsomal versus reductase of the mitochondrial P450 systems are completely different and show no homology.[24]
Redox
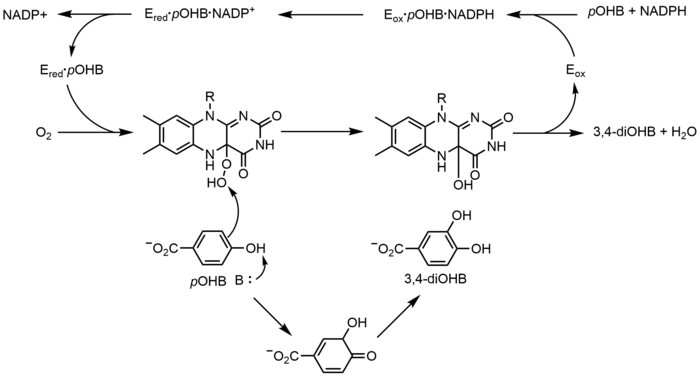
p-Hydroxybenzoate hydroxylase (PHBH) catalyzes the oxygenation of p-hydroxybenzoate (pOHB) to 3,4-dihyroxybenzoate (3,4-diOHB); FAD, NADPH and molecular oxygen are all required for this reaction. NADPH first transfers a hydride equivalent to FAD, creating FADH−, and then NADP+ dissociates from the enzyme. Reduced PHBH then reacts with molecular oxygen to form the flavin-C(4a)-hydroperoxide. The flavin hydroperoxide quickly hydroxylates pOHB, and then eliminates water to regenerate oxidized flavin.[23] An alternative flavin-mediated oxygenation mechanism involves the use of a flavin-N(5)-oxide rather than a flavin-C(4a)-(hydro)peroxide.[2][3]
Nonredox
Chorismate synthase (CS) catalyzes the last step in the shikimate pathway—the formation of chorismate. Two classes of CS are known, both of which require FMN, but are divided on their need for NADPH as a reducing agent. The proposed mechanism for CS involves radical species. The radical flavin species has not been detected spectroscopically without using a substrate analogue, which suggests that it is short-lived. However, when using a fluorinated substrate, a neutral flavin semiquinone was detected.[23]
Complex flavoenzymes
Glutamate synthase catalyzes the conversion of 2-oxoglutarate into L-glutamate with L-glutamine serving as the nitrogen source for the reaction. All glutamate syntheses are iron-sulfur flavoproteins containing an iron-sulfur cluster and FMN. The three classes of glutamate syntheses are categorized based on their sequences and biochemical properties. Even though there are three classes of this enzyme, it is believed that they all operate through the same mechanism, only differing by what first reduces the FMN. The enzyme produces two glutamate molecules: one by the hydrolysis of glutamine (forming glutamate and ammonia), and the second by the ammonia produced from the first reaction attacking 2-oxoglutarate, which is reduced by FMN to glutamate.[23]
Clinical significance
Flavoprotein-related diseases
Due to the importance of flavoproteins, it is unsurprising that approximately 60% of human flavoproteins cause human disease when mutated.[17] In some cases, this is due to a decreased affinity for FAD or FMN and so excess riboflavin intake may lessen disease symptoms, such as for multiple acyl-CoA dehydrogenase deficiency.[9] In addition, riboflavin deficiency itself (and the resulting lack of FAD and FMN) can cause health issues.[9] For example, in ALS patients, there are decreased levels of FAD synthesis.[9] Both of these paths can result in a variety of symptoms, including developmental or gastrointestinal abnormalities, faulty fat break-down, anemia, neurological problems, cancer or heart disease, migraine, worsened vision and skin lesions.[9] The pharmaceutical industry therefore produces riboflavin to supplement diet in certain cases. In 2008, the global need for riboflavin was 6,000 tons per year, with production capacity of 10,000 tons.[4] This $150 to 500 million market is not only for medical applications, but is also used as a supplement to animal food in the agricultural industry and as a food colorant.[4]
Drug design
New design of anti-bacterial medications is of continuing importance in scientific research as bacterial antibiotic resistance to common antibiotics increases. A specific metabolic protein that uses FAD (Complex II) is vital for bacterial virulence, and so targeting FAD synthesis or creating FAD analogs could be a useful area of investigation.[28] Already, scientists have determined the two structures FAD usually assumes once bound: either an extended or a butterfly conformation, in which the molecule essentially folds in half, resulting in the stacking of the adenine and isoalloxazine rings.[14] FAD imitators that are able to bind in a similar manner but do not permit protein function could be useful mechanisms of inhibiting bacterial infection.[14] Alternatively, drugs blocking FAD synthesis could achieve the same goal; this is especially intriguing because human and bacterial FAD synthesis relies on very different enzymes, meaning that a drug made to target bacterial FAD synthase would be unlikely to interfere with the human FAD synthase enzymes.[29]
Optogenetics
Optogenetics allows control of biological events in a non-invasive manner.[30] The field has advanced in recent years with a number of new tools, including those to trigger light sensitivity, such as the Blue-Light-Utilizing FAD domains (BLUF). BLUFs encode a 100 to 140 amino acid sequence that was derived from photoreceptors in plants and bacteria.[30] Similar to other photoreceptors, the light causes structural changes in the BLUF domain that results in disruption of downstream interactions.[30] Current research investigates proteins with the appended BLUF domain and how different external factors can impact the proteins.[30]
Treatment monitoring
There are a number of molecules in the body that have native fluorescence including tryptophan, collagen, FAD, NADH and porphyrins.[31] Scientists have taken advantage of this by using them to monitor disease progression or treatment effectiveness or aid in diagnosis. For instance, native fluorescence of a FAD and NADH is varied in normal tissue and oral submucous fibrosis, which is an early sign of invasive oral cancer.[31] Doctors therefore have been employing fluorescence to assist in diagnosis and monitor treatment as opposed to the standard biopsy.[31]
Additional images
- FADH2
See also
References
- ^ Teufel, Robin; Agarwal, Vinayak; Moore, Bradley S. (2016-04-01). "Unusual flavoenzyme catalysis in marine bacteria". Current Opinion in Chemical Biology. 31: 31–39. doi:10.1016/j.cbpa.2016.01.001. ISSN 1879-0402. PMC 4870101. PMID 26803009.
- ^ a b Teufel, R; Miyanaga, A; Michaudel, Q; Stull, F; Louie, G; Noel, JP; Baran, PS; Palfey, B; Moore, BS (28 November 2013). "Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement". Nature. 503 (7477): 552–6. Bibcode:2013Natur.503..552T. doi:10.1038/nature12643. PMC 3844076. PMID 24162851.
- ^ a b Teufel, Robin; Stull, Frederick; Meehan, Michael J.; Michaudel, Quentin; Dorrestein, Pieter C.; Palfey, Bruce; Moore, Bradley S. (2015-07-01). "Biochemical Establishment and Characterization of EncM's Flavin-N5-oxide Cofactor". Journal of the American Chemical Society. 137 (25): 8078–8085. doi:10.1021/jacs.5b03983. ISSN 1520-5126. PMC 4720136. PMID 26067765.
- ^ a b c Abbas CA, Sibirny AA (Jun 2011). "Genetic control of biosynthesis and transport of riboflavin and flavin nucleotides and construction of robust biotechnological producers". Microbiology and Molecular Biology Reviews. 75 (2): 321–60. doi:10.1128/mmbr.00030-10. PMC 3122625. PMID 21646432.
- ^ Hayashi H (2013). B Vitamins and Folate: Chemistry, Analysis, Function and Effects. Cambridge, UK: The Royal Society of Chemistry. p. 7. ISBN 978-1-84973-369-4.
- ^ Warburg O, Christian W (1938). "Isolation of the prosthetic group of the amino acid oxidase". Biochemische Zeitschrift. 298: 150–168.
- ^ Metzler DE, Metzler CM, Sauke DJ (2003). Biochemistry (2nd ed.). San Diego: Harcourt, Academic Press. ISBN 978-0-12-492541-0.
- ^ a b Devlin TM (2011). Textbook of Biochemistry: with Clinical Correlations (7th ed.). Hoboken, NJ: John Wiley & Sons. ISBN 978-0-470-28173-4.
- ^ a b c d e f g h i j k Barile M, Giancaspero TA, Brizio C, Panebianco C, Indiveri C, Galluccio M, Vergani L, Eberini I, Gianazza E (2013). "Biosynthesis of flavin cofactors in man: implications in health and disease". Current Pharmaceutical Design. 19 (14): 2649–75. doi:10.2174/1381612811319140014. PMID 23116402.
- ^ Teufel, Robin; Miyanaga, Akimasa; Michaudel, Quentin; Stull, Frederick; Louie, Gordon; Noel, Joseph P.; Baran, Phil S.; Palfey, Bruce; Moore, Bradley S. (2013-11-28). "Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement". Nature. 503 (7477): 552–556. Bibcode:2013Natur.503..552T. doi:10.1038/nature12643. ISSN 1476-4687. PMC 3844076. PMID 24162851.
- ^ a b c d e f Kim HJ, Winge DR (May 2013). "Emerging concepts in the flavinylation of succinate dehydrogenase". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1827 (5): 627–36. doi:10.1016/j.bbabio.2013.01.012. PMC 3626088. PMID 23380393.
- ^ Liu S (2012). Bioprocess Engineering: Kinetics, Sustainability, and Reactor Design. Newnes. ISBN 978-0-444-63783-3.
- ^ Lewis JA, Escalante-Semerena JC (Aug 2006). "The FAD-dependent tricarballylate dehydrogenase (TcuA) enzyme of Salmonella enterica converts tricarballylate into cis-aconitate". Journal of Bacteriology. 188 (15): 5479–86. doi:10.1128/jb.00514-06. PMC 1540016. PMID 16855237.
- ^ a b c Kuppuraj G, Kruise D, Yura K (Nov 2014). "Conformational behavior of flavin adenine dinucleotide: conserved stereochemistry in bound and free states". The Journal of Physical Chemistry B. 118 (47): 13486–97. doi:10.1021/jp507629n. PMID 25389798.
- ^ Monteira M (2013). B Vitamins and Folate: Chemistry, Analysis, Function and Effects. Cambridge, UK: The Royal Society of Chemistry. p. 94. ISBN 978-1-84973-369-4.
- ^ a b c Macheroux P, Kappes B, Ealick SE (Aug 2011). "Flavogenomics--a genomic and structural view of flavin-dependent proteins". The FEBS Journal. 278 (15): 2625–34. doi:10.1111/j.1742-4658.2011.08202.x. PMID 21635694. S2CID 22220250.
- ^ a b c d Lienhart WD, Gudipati V, Macheroux P (Jul 2013). "The human flavoproteome". Archives of Biochemistry and Biophysics. 535 (2): 150–62. doi:10.1016/j.abb.2013.02.015. PMC 3684772. PMID 23500531.
- ^ Hühner J, Ingles-Prieto Á, Neusüß C, Lämmerhofer M, Janovjak H (Feb 2015). "Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in mammalian model cells by CE with LED-induced fluorescence detection". Electrophoresis. 36 (4): 518–25. doi:10.1002/elps.201400451. PMID 25488801. S2CID 27285540.
- ^ Stryer L, Berg JM, Tymoczko JL (2007). Biochemistry (6th ed.). New York: Freeman. ISBN 978-0-7167-8724-2.
- ^ a b c Mansoorabadi SO, Thibodeaux CJ, Liu HW (Aug 2007). "The diverse roles of flavin coenzymes--nature's most versatile thespians". The Journal of Organic Chemistry. 72 (17): 6329–42. doi:10.1021/jo0703092. PMC 2519020. PMID 17580897.
- ^ King MW (18 May 2020). "Vitamins, Minerals, Supplements". The Medical Biochemistry Page.
- ^ Garma, Leonardo D.; Medina, Milagros; Juffer, André H. (2016-11-01). "Structure-based classification of FAD binding sites: A comparative study of structural alignment tools". Proteins: Structure, Function, and Bioinformatics. 84 (11): 1728–1747. doi:10.1002/prot.25158. ISSN 1097-0134. PMID 27580869. S2CID 26066208.
- ^ a b c d e f g h Fagan RL, Palfey BA (2010). "Flavin-Dependent Enzymes". Comprehensive Natural Products II Chemistry and Biology. 7: 37–113.
- ^ a b Hanukoglu I (1996). "Electron transfer proteins of cytochrome P450 systems" (PDF). Adv. Mol. Cell Biol. Advances in Molecular and Cell Biology. 14: 29–55. doi:10.1016/S1569-2558(08)60339-2. ISBN 9780762301133.
- ^ Ziegler GA, Vonrhein C, Hanukoglu I, Schulz GE (Jun 1999). "The structure of adrenodoxin reductase of mitochondrial P450 systems: electron transfer for steroid biosynthesis". Journal of Molecular Biology. 289 (4): 981–90. doi:10.1006/jmbi.1999.2807. PMID 10369776.
- ^ a b Hanukoglu I (2017). "Conservation of the Enzyme-Coenzyme Interfaces in FAD and NADP Binding Adrenodoxin Reductase-A Ubiquitous Enzyme". Journal of Molecular Evolution. 85 (5): 205–218. Bibcode:2017JMolE..85..205H. doi:10.1007/s00239-017-9821-9. PMID 29177972. S2CID 7120148.
- ^ Hanukoglu I, Jefcoate CR (Apr 1980). "Mitochondrial cytochrome P-450scc. Mechanism of electron transport by adrenodoxin" (PDF). The Journal of Biological Chemistry. 255 (7): 3057–61. doi:10.1016/S0021-9258(19)85851-9. PMID 6766943.
- ^ McNeil MB, Fineran PC (May 2013). "Prokaryotic assembly factors for the attachment of flavin to complex II". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1827 (5): 637–47. doi:10.1016/j.bbabio.2012.09.003. PMID 22985599.
- ^ Serrano A, Ferreira P, Martínez-Júlvez M, Medina M (2013). "The prokaryotic FAD synthetase family: a potential drug target". Current Pharmaceutical Design. 19 (14): 2637–48. doi:10.2174/1381612811319140013. PMID 23116401.
- ^ a b c d Christie JM, Gawthorne J, Young G, Fraser NJ, Roe AJ (May 2012). "LOV to BLUF: flavoprotein contributions to the optogenetic toolkit". Molecular Plant. 5 (3): 533–44. doi:10.1093/mp/sss020. PMID 22431563.
- ^ a b c Sivabalan S, Vedeswari CP, Jayachandran S, Koteeswaran D, Pravda C, Aruna PR, Ganesan S (2010). "In vivo native fluorescence spectroscopy and nicotinamide adinine dinucleotide/flavin adenine dinucleotide reduction and oxidation states of oral submucous fibrosis for chemopreventive drug monitoring". Journal of Biomedical Optics. 15 (1): 017010–017010–11. Bibcode:2010JBO....15a7010S. doi:10.1117/1.3324771. PMID 20210484. S2CID 40028193.
External links
- FAD bound to proteins in the PDB
- FAD entry in the NIH Chemical Database