Cis-regulatory element
Cis-regulatory elements (CREs) or cis-regulatory modules (CRMs) are regions of non-coding DNA which regulate the transcription of neighboring genes. CREs are vital components of genetic regulatory networks, which in turn control morphogenesis, the development of anatomy, and other aspects of embryonic development, studied in evolutionary developmental biology.
CREs are found in the vicinity of the genes that they regulate. CREs typically regulate gene transcription by binding to transcription factors. A single transcription factor may bind to many CREs, and hence control the expression of many genes (pleiotropy). The Latin prefix cis means "on this side", i.e. on the same molecule of DNA as the gene(s) to be transcribed.
CRMs are stretches of DNA, usually 100–1000 DNA base pairs in length,[1] where a number of transcription factors can bind and regulate expression of nearby genes and regulate their transcription rates. They are labeled as cis because they are typically located on the same DNA strand as the genes they control as opposed to trans, which refers to effects on genes not located on the same strand or farther away, such as transcription factors.[1] One cis-regulatory element can regulate several genes,[2] and conversely, one gene can have several cis-regulatory modules.[3] Cis-regulatory modules carry out their function by integrating the active transcription factors and the associated co-factors at a specific time and place in the cell where this information is read and an output is given.[4]
CREs are often but not always upstream of the transcription site. CREs contrast with trans-regulatory elements (TREs). TREs code for transcription factors.[citation needed]
Overview
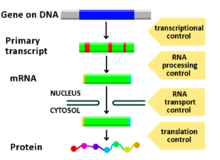
The genome of an organism contains anywhere from a few hundred to thousands of different genes, all encoding a singular product or more. For numerous reasons, including organizational maintenance, energy conservation, and generating phenotypic variance, it is important that genes are only expressed when they are needed. The most efficient way for an organism to regulate gene expression is at the transcriptional level. CREs function to control transcription by acting nearby or within a gene. The most well characterized types of CREs are enhancers and promoters. Both of these sequence elements are structural regions of DNA that serve as transcriptional regulators.[citation needed]
Cis-regulatory modules are one of several types of functional regulatory elements. Regulatory elements are binding sites for transcription factors, which are involved in gene regulation.[1] Cis-regulatory modules perform a large amount of developmental information processing.[1] Cis-regulatory modules are non-random clusters at their specified target site that contain transcription factor binding sites.[1]
The original definition presented cis-regulatory modules as enhancers of cis-acting DNA, which increased the rate of transcription from a linked promoter.[4] However, this definition has changed to define cis-regulatory modules as a DNA sequence with transcription factor binding sites which are clustered into modular structures, including -but not limited to- locus control regions, promoters, enhancers, silencers, boundary control elements and other modulators.[4]
Cis-regulatory modules can be divided into three classes; enhancers, which regulate gene expression positively;[1] insulators, which work indirectly by interacting with other nearby cis-regulatory modules; and[1] silencers that turn off expression of genes.[1]
The design of cis-regulatory modules is such that transcription factors and epigenetic modifications serve as inputs, and the output of the module is the command given to the transcription machinery, which in turn determines the rate of gene transcription or whether it is turned on or off.[1] There are two types of transcription factor inputs: those that determine when the target gene is to be expressed and those that serve as functional drivers, which come into play only during specific situations during development.[1] These inputs can come from different time points, can represent different signal ligands, or can come from different domains or lineages of cells. However, a lot still remains unknown.[citation needed]
Additionally, the regulation of chromatin structure and nuclear organization also play a role in determining and controlling the function of cis-regulatory modules.[4] Thus gene-regulation functions (GRF) provide a unique characteristic of a cis-regulatory module (CRM), relating the concentrations of transcription factors (input) to the promoter activities (output). The challenge is to predict GRFs. This challenge still remains unsolved. In general, gene-regulation functions do not use Boolean logic,[2] although in some cases the approximation of the Boolean logic is still very useful.[citation needed]
The Boolean logic assumption
Within the assumption of the Boolean logic, principles guiding the operation of these modules includes the design of the module which determines the regulatory function. In relation to development, these modules can generate both positive and negative outputs. The output of each module is a product of the various operations performed on it. Common operations include the OR gate – this design indicates that in an output will be given when either input is given [3], and the AND gate – in this design two different regulatory factors are necessary to make sure that a positive output results.[1] "Toggle Switches" – This design occurs when the signal ligand is absent while the transcription factor is present; this transcription factor ends up acting as a dominant repressor. However, once the signal ligand is present the transcription factor's role as repressor is eliminated and transcription can occur.[1]
Other Boolean logic operations can occur as well, such as sequence specific transcriptional repressors, which when they bind to the cis-regulatory module lead to an output of zero. Additionally, besides influence from the different logic operations, the output of a "cis"-regulatory module will also be influenced by prior events.[1] 4) Cis-regulatory modules must interact with other regulatory elements. For the most part, even with the presence of functional overlap between cis-regulatory modules of a gene, the modules' inputs and outputs tend to not be the same.[1]
While the assumption of Boolean logic is important for systems biology, detailed studies show that in general the logic of gene regulation is not Boolean.[2] This means, for example, that in the case of a cis-regulatory module regulated by two transcription factors, experimentally determined gene-regulation functions can not be described by the 16 possible Boolean functions of two variables. Non-Boolean extensions of the gene-regulatory logic have been proposed to correct for this issue.[2]
Classification
Cis-regulatory modules can be characterized by the information processing that they encode and the organization of their transcription factor binding sites. Additionally, cis-regulatory modules are also characterized by the way they affect the probability, proportion, and rate of transcription.[4] Highly cooperative and coordinated cis-regulatory modules are classified as enhanceosomes.[4] The architecture and the arrangement of the transcription factor binding sites are critical because disruption of the arrangement could cancel out the function.[4] Functional flexible cis-regulatory modules are called billboards. Their transcriptional output is the summation effect of the bound transcription factors.[4] Enhancers affect the probability of a gene being activated, but have little or no effect on rate.[4] The Binary response model acts like an on/off switch for transcription. This model will increase or decrease the amount of cells that transcribe a gene, but it does not affect the rate of transcription.[4] Rheostatic response model describes cis-regulatory modules as regulators of the initiation rate of transcription of its associated gene.[4]
Promoter
Promoters are CREs consisting of relatively short sequences of DNA which include the site where transcription is initiated and the region approximately 35 bp upstream or downstream from the initiation site (bp).[5] In eukaryotes, promoters usually have the following four components: the TATA box, a TFIIB recognition site, an initiator, and the downstream core promoter element.[5] It has been found that a single gene can contain multiple promoter sites.[6] In order to initiate transcription of the downstream gene, a host of DNA-binding proteins called transcription factors (TFs) must bind sequentially to this region.[5] Only once this region has been bound with the appropriate set of TFs, and in the proper order, can RNA polymerase bind and begin transcribing the gene.
Enhancers
Enhancers are CREs that influence (enhance) the transcription of genes on the same molecule of DNA and can be found upstream, downstream, within the introns, or even relatively far away from the gene they regulate. Multiple enhancers can act in a coordinated fashion to regulate transcription of one gene.[7] A number of genome-wide sequencing projects have revealed that enhancers are often transcribed to long non-coding RNA (lncRNA) or enhancer RNA (eRNA), whose changes in levels frequently correlate with those of the target gene mRNA.[8]
Silencers
Silencers are CREs that can bind transcription regulation factors (proteins) called repressors, thereby preventing transcription of a gene. The term "silencer" can also refer to a region in the 3' untranslated region of messenger RNA, that binds proteins which suppress translation of that mRNA molecule, but this usage is distinct from its use in describing a CRE.[citation needed]
Operators
Operators are CREs in prokaryotes and some eukaryotes that exist within operons, where they can bind proteins called repressors to affect transcription.[citation needed]
Evolutionary role
CREs have an important evolutionary role. The coding regions of genes are often well conserved among organisms; yet different organisms display marked phenotypic diversity. It has been found that polymorphisms occurring within non-coding sequences have a profound effect on phenotype by altering gene expression.[7] Mutations arising within a CRE can generate expression variance by changing the way TFs bind. Tighter or looser binding of regulatory proteins will lead to up- or down-regulated transcription.
Cis-regulatory module in gene regulatory network
The function of a gene regulatory network depends on the architecture of the nodes, whose function is dependent on the multiple cis-regulatory modules.[1] The layout of cis-regulatory modules can provide enough information to generate spatial and temporal patterns of gene expression.[1] During development each domain, where each domain represents a different spatial regions of the embryo, of gene expression will be under the control of different cis-regulatory modules.[1] The design of regulatory modules help in producing feedback, feed forward, and cross-regulatory loops.[9]
Mode of action
Cis-regulatory modules can regulate their target genes over large distances. Several models have been proposed to describe the way that these modules may communicate with their target gene promoter.[4] These include the DNA scanning model, the DNA sequence looping model and the facilitated tracking model. In the DNA scanning model, the transcription factor and cofactor complex form at the cis-regulatory module and then continues to move along the DNA sequence until it finds the target gene promoter.[4] In the looping model, the transcription factor binds to the cis-regulatory module, which then causes the looping of the DNA sequence and allows for the interaction with the target gene promoter. The transcription factor-cis-regulatory module complex causes the looping of the DNA sequence slowly towards the target promoter and forms a stable looped configuration.[4] The facilitated tracking model combines parts of the two previous models.
Identification and computational prediction
Besides experimentally determining CRMs, there are various bioinformatics algorithms for predicting them. Most algorithms try to search for significant combinations of transcription factor binding sites (DNA binding sites) in promoter sequences of co-expressed genes.[10] More advanced methods combine the search for significant motifs with correlation in gene expression datasets between transcription factors and target genes.[11] Both methods have been implemented, for example, in the ModuleMaster. Other programs created for the identification and prediction of cis-regulatory modules include:
INSECT 2.0[12] is a web server that allows to search Cis-regulatory modules in a genome-wide manner. The program relies on the definition of strict restrictions among the Transcription Factor Binding Sites (TFBSs) that compose the module in order to decrease the false positives rate. INSECT is designed to be user-friendly since it allows automatic retrieval of sequences and several visualizations and links to third-party tools in order to help users to find those instances that are more likely to be true regulatory sites. INSECT 2.0 algorithm was previously published and the algorithm and theory behind it explained in[13]
Stubb uses hidden Markov models to identify statistically significant clusters of transcription factor combinations. It also uses a second related genome to improve the prediction accuracy of the model.[14]
Bayesian Networks use an algorithm that combines site predictions and tissue-specific expression data for transcription factors and target genes of interest. This model also uses regression trees to depict the relationship between the identified cis-regulatory module and the possible binding set of transcription factors.[15]
CRÈME examine clusters of target sites for transcription factors of interest. This program uses a database of confirmed transcription factor binding sites that were annotated across the human genome. A search algorithm is applied to the data set to identify possible combinations of transcription factors, which have binding sites that are close to the promoter of the gene set of interest. The possible cis-regulatory modules are then statistically analyzed and the significant combinations are graphically represented[16]
Active cis-regulatory modules in a genomic sequence have been difficult to identify. Problems in identification arise because often scientists find themselves with a small set of known transcription factors, so it makes it harder to identify statistically significant clusters of transcription factor binding sites.[14] Additionally, high costs limit the use of large whole genome tiling arrays.[15]

Examples
An example of a cis-acting regulatory sequence is the operator in the lac operon. This DNA sequence is bound by the lac repressor, which, in turn, prevents transcription of the adjacent genes on the same DNA molecule. The lac operator is, thus, considered to "act in cis" on the regulation of the nearby genes. The operator itself does not code for any protein or RNA.
In contrast, trans-regulatory elements are diffusible factors, usually proteins, that may modify the expression of genes distant from the gene that was originally transcribed to create them. For example, a transcription factor that regulates a gene on chromosome 6 might itself have been transcribed from a gene on chromosome 11. The term trans-regulatory is constructed from the Latin root trans, which means "across from".
There are cis-regulatory and trans-regulatory elements. Cis-regulatory elements are often binding sites for one or more trans-acting factors.
To summarize, cis-regulatory elements are present on the same molecule of DNA as the gene they regulate whereas trans-regulatory elements can regulate genes distant from the gene from which they were transcribed.
Examples in RNA
| Type | Abbr. | Function | Distribution | Ref. |
|---|---|---|---|---|
| Frameshift element | Regulates alternative frame use with messenger RNAs | Archaea, bacteria, Eukaryota, RNA viruses | [17][18][19] | |
| Internal ribosome entry site | IRES | Initiates translation in the middle of a messenger RNA | RNA virus, Eukaryota | [20] |
| Iron response element | IRE | Regulates the expression of iron associated genes | Eukaryota | [21] |
| Leader peptide | Regulates transcription of associated genes and/or operons | Bacteria | [22] | |
| Riboswitch | Gene regulation | Bacteria, Eukaryota | [23] | |
| RNA thermometer | Gene regulation | Bacteria | [24] | |
| Selenocysteine insertion sequence | SECIS | Directs the cell to translate UGA stop-codons as selenocysteines | Metazoa | [25] |
See also
- DNA
- RNA
- List of cis-regulatory RNA elements
- Polyadenylation signals, mRNA
- AU-rich element, mRNA
- Other
References
- ^ a b c d e f g h i j k l m n o p q Davidson EH (2006). The Regulatory Genome: Gene Regulatory Networks in Development and Evolution. Elsevier. pp. 1–86.
- ^ a b c d Teif V.B. (2010). "Predicting Gene-Regulation Functions: Lessons from Temperate Bacteriophages". Biophysical Journal. 98 (7): 1247–56. Bibcode:2010BpJ....98.1247T. doi:10.1016/j.bpj.2009.11.046. PMC 2849075. PMID 20371324.
- ^ Ben-Tabou de-Leon S, Davidson EH (2007). "Gene regulation: gene control network in development" (PDF). Annu Rev Biophys Biomol Struct. 36: 191–212. doi:10.1146/annurev.biophys.35.040405.102002. PMID 17291181.
- ^ a b c d e f g h i j k l m n Jeziorska DM, Jordan KW, Vance KW (2009). "A systems biology approach to understanding cis-regulatory module function". Semin. Cell Dev. Biol. 20 (7): 856–862. doi:10.1016/j.semcdb.2009.07.007. PMID 19660565.
- ^ a b c Butler JE, Kadonaga JT (October 2002). "The RNA polymerase II core promoter: a key component in the regulation of gene expression". Genes & Development. 16 (20): 2583–2592. doi:10.1101/gad.1026202. PMID 12381658.
- ^ Choi S (17 May 2008). Introduction to Systems Biology. Springer Science & Business Media. p. 78. ISBN 978-1-59745-531-2.
- ^ a b Wittkopp PJ, Kalay G (December 2011). "Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence". Nature Reviews Genetics. 13 (1): 59–69. doi:10.1038/nrg3095. PMID 22143240. S2CID 13513643.
- ^ Melamed P, Yosefzun Y, et al. (2 March 2016). "Transcriptional enhancers: Transcription, function and flexibility". Transcription. 7 (1): 26–31. doi:10.1080/21541264.2015.1128517. PMC 4802784. PMID 26934309.
- ^ Li E, Davidson EH (2009). "Building Developmental Gene Regulatory Networks". Birth Defects Res. 87 (2): 123–130. doi:10.1002/bdrc.20152. PMC 2747644. PMID 19530131.
- ^ Aerts, S.; et al. (2003). "Computational detection of cis-regulatory modules". Bioinformatics. 19 (Suppl 2): ii5–14. doi:10.1093/bioinformatics/btg1052. PMID 14534164.
- ^ Wrzodek, Clemens; Schröder, Adrian; Dräger, Andreas; Wanke, Dierk; Berendzen, Kenneth W.; Kronfeld, Marcel; Harter, Klaus; Zell, Andreas (2010). "ModuleMaster: A new tool to decipher transcriptional regulatory networks". Biosystems. 99 (1). Ireland: Elsevier: 79–81. doi:10.1016/j.biosystems.2009.09.005. ISSN 0303-2647. PMID 19819296.
- ^ Parra RG, Rohr CO, Koile D, Perez-Castro C, Yankilevich P (2015). "INSECT 2.0: a web-server for genome-wide cis-regulatory modules prediction". Bioinformatics. 32 (8): 1229–31. doi:10.1093/bioinformatics/btv726. hdl:11336/37980. PMID 26656931.
- ^ Rohr CO, Parra RG, Yankilevich P, Perez-Castro C (2013). "INSECT: IN-silico SEarch for Co-occurring Transcription factors". Bioinformatics. 29 (22): 2852–8. doi:10.1093/bioinformatics/btt506. hdl:11336/12301. PMID 24008418.
- ^ a b Sinha S, Liang Y, Siggia E (2006). "Stubb: a program for discovery and analysis of cis-regulatory modules". Nucleic Acids Res. 34 (Web Server issue): W555–W559. doi:10.1093/nar/gkl224. PMC 1538799. PMID 16845069.
- ^ a b Chen X, Blanchette M (2007). "Comparing sequences without using alignments: application to HIV/SIV subtyping". BMC Bioinformatics. 8: 1–17. doi:10.1186/1471-2105-8-1. PMC 1766362. PMID 17199892.
- ^ Sharan R, Ben-Hur A, Loots GG, Ovcharenko I (2004). "CREME: Cis-Regulatory Module Explorer for the human genome". Nucleic Acids Res. 32 (Web Server issue): W253–W256. doi:10.1093/nar/gkh385. PMC 441523. PMID 15215390.
- ^ Bekaert M, Firth AE, Zhang Y, Gladyshev VN, Atkins JF, Baranov PV (January 2010). "Recode-2: new design, new search tools, and many more genes". Nucleic Acids Research. 38 (Database issue): D69–74. doi:10.1093/nar/gkp788. PMC 2808893. PMID 19783826.
- ^ Chung BY, Firth AE, Atkins JF (March 2010). "Frameshifting in alphaviruses: a diversity of 3' stimulatory structures". Journal of Molecular Biology. 397 (2): 448–456. doi:10.1016/j.jmb.2010.01.044. PMID 20114053.
- ^ Giedroc DP, Cornish PV (February 2009). "Frameshifting RNA pseudoknots: structure and mechanism". Virus Research. 139 (2): 193–208. doi:10.1016/j.virusres.2008.06.008. PMC 2670756. PMID 18621088.
- ^ Mokrejs M, Vopálenský V, Kolenaty O, Masek T, Feketová Z, Sekyrová P, Skaloudová B, Kríz V, Pospísek M (January 2006). "IRESite: the database of experimentally verified IRES structures (www.iresite.org)". Nucleic Acids Research. 34 (Database issue): D125–130. doi:10.1093/nar/gkj081. PMC 1347444. PMID 16381829.
- ^ Hentze MW, Kühn LC (August 1996). "Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide, and oxidative stress". Proceedings of the National Academy of Sciences of the United States of America. 93 (16): 8175–8182. Bibcode:1996PNAS...93.8175H. doi:10.1073/pnas.93.16.8175. PMC 38642. PMID 8710843.
- ^ Platt T (1986). "Transcription termination and the regulation of gene expression". Annual Review of Biochemistry. 55: 339–372. doi:10.1146/annurev.bi.55.070186.002011. PMID 3527045.
- ^ Breaker RR (March 2008). "Complex riboswitches". Science. 319 (5871): 1795–1797. Bibcode:2008Sci...319.1795B. doi:10.1126/science.1152621. PMID 18369140. S2CID 45588146.
- ^ Kortmann J, Narberhaus F (March 2012). "Bacterial RNA thermometers: molecular zippers and switches". Nature Reviews. Microbiology. 10 (4): 255–265. doi:10.1038/nrmicro2730. PMID 22421878. S2CID 29414695.
- ^ Walczak R, Westhof E, Carbon P, Krol A (April 1996). "A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs". RNA. 2 (4): 367–379. PMC 1369379. PMID 8634917.
Further reading
- Wray GA (March 2007). "The evolutionary significance of cis-regulatory mutations". Nature Reviews Genetics. 8 (3): 206–216. doi:10.1038/nrg2063. PMID 17304246. S2CID 560067.
- Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB (February 2005). "Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila". Nature. 433 (7025): 481–487. Bibcode:2005Natur.433..481G. doi:10.1038/nature03235. PMID 15690032. S2CID 16422483.
- Prud'homme B, Gompel N, Rokas A, Kassner VA, Williams TM, Yeh SD, True JR, Carroll SB (April 2006). "Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene". Nature. 440 (7087): 1050–1053. Bibcode:2006Natur.440.1050P. doi:10.1038/nature04597. PMID 16625197. S2CID 9581516.
- Stern DL (August 2000). "Evolutionary developmental biology and the problem of variation". Evolution; International Journal of Organic Evolution. 54 (4): 1079–1091. doi:10.1111/j.0014-3820.2000.tb00544.x. PMID 11005278.
- Weatherbee SD, Carroll SB, Grenier JK (2004). From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. Cambridge, MA: Blackwell Publishers. ISBN 978-1-4051-1950-4.
External links
- Gene Regulation Info – manually curated lists of resources, reviews, community discussions
- Cellular Darwinism
- Regulation of Gene Expression at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
