COLD-PCR
COLD-PCR (co-amplification at lower denaturation temperature PCR) is a modified polymerase chain reaction (PCR) protocol that enriches variant alleles from a mixture of wildtype and mutation-containing DNA. The ability to preferentially amplify and identify minority alleles and low-level somatic DNA mutations in the presence of excess wildtype alleles is useful for the detection of mutations. Detection of mutations is important in the case of early cancer detection from tissue biopsies and body fluids such as blood plasma or serum, assessment of residual disease after surgery or chemotherapy, disease staging and molecular profiling for prognosis or tailoring therapy to individual patients, and monitoring of therapy outcome and cancer remission or relapse. Common PCR will amplify both the major (wildtype) and minor (mutant) alleles with the same efficiency, occluding the ability to easily detect the presence of low-level mutations. The capacity to detect a mutation in a mixture of variant/wildtype DNA is valuable because this mixture of variant DNAs can occur when provided with a heterogeneous sample – as is often the case with cancer biopsies. Currently, traditional PCR is used in tandem with a number of different downstream assays for genotyping or the detection of somatic mutations. These can include the use of amplified DNA for RFLP analysis, MALDI-TOF (matrix-assisted laser-desorption–time-of-flight) genotyping, or direct sequencing for detection of mutations by Sanger sequencing or pyrosequencing. Replacing traditional PCR with COLD-PCR for these downstream assays will increase the reliability in detecting mutations from mixed samples, including tumors and body fluids.
Method overviews
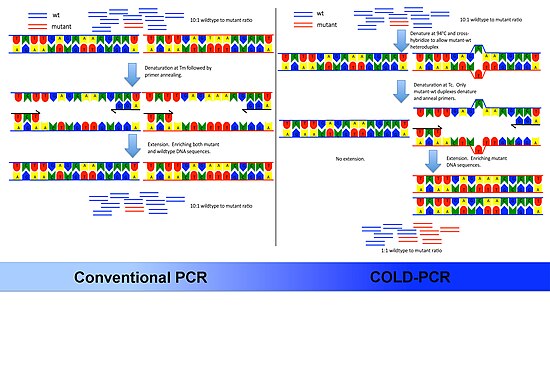
The underlying principle of COLD-PCR is that single nucleotide mismatches will slightly alter the melting temperature (Tm) of the double-stranded DNA. Depending on the sequence context and position of the mismatch, Tm changes of 0.2–1.5 °C (0.36–2.7 °F) are common for sequences up to 200bp or higher. Knowing this the authors of the protocol took advantage of two observations:
- Each double-stranded DNA has a 'critical temperature' (Tc) lower than its Tm. The PCR amplification efficiency drops measurably below the Tc.
- The Tc is dependent on DNA sequence. Two template DNA fragments differing by only one or two nucleotide mismatches will have different amplification efficiencies if the denaturation step of PCR is set to the Tc.
Keeping these principles in mind the authors developed the following general protocol:
- Denaturation stage. DNA is denatured at a high temperature – usually 94 °C (201 °F).
- Intermediate annealing stage. Set an intermediate annealing temperature that allows hybridization of mutant and wildtype allele DNA to one another. Because the mutant allele DNA forms the minority of DNA in the mixture they will be more likely to form mismatch heteroduplex DNA with the wildtype DNA.
- Melting stage. These heteroduplexes will more readily melt at lower temperatures. Hence they are selectively denatured at the Tc.
- Primer annealing stage. The homo-duplex DNA will preferentially remain double stranded and not be available for primer annealing.
- Extension stage. The DNA polymerase will extend complementary to the template DNA. Since the heteroduplex DNA is used as template, a larger proportion of minor variant DNA will be amplified and be available for subsequent rounds of PCR.
There are two forms of COLD-PCR that have been developed to date. Full COLD-PCR and fast COLD-PCR.
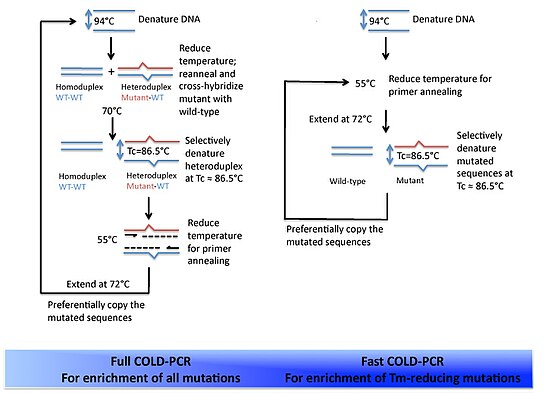
Full
Full COLD-PCR is identical to the protocol outlined above. These five stages are used for each round of amplification.
Fast
Fast COLD-PCR differs from Full COLD-PCR in that the denaturation and intermediate annealing stages are skipped. This is because, in some cases, the preferential amplification of the mutant DNA is so great that ensuring the formation of the mutant/wildtype heteroduplex DNA is not needed. Thus the denaturation can occur at the Tc, proceed to primer annealing, and then polymerase-mediated extension. Each round of amplification will include these three stages in that order. By utilizing the lower denaturation temperature, the reaction will discriminate toward the products with the lower Tm – i.e. the variant alleles. Fast COLD-PCR produces much faster results due to the shortened protocol, while Full COLD-PCR is essential for amplification of all possible mutations in the starting mixture of DNA.
Two-round COLD-PCR is a modified version of Fast COLD-PCR. During the second round of Fast COLD-PCR nested primers are used. This improves the sensitivity of mutation detection compared to one-round Fast COLD-PCR.[1]
Uses
COLD-PCR has been used to improve the reliability of a number of different assays that traditionally use conventional PCR.

RFLP
A restriction fragment length polymorphism results in the cleavage (or absence thereof) of DNA for a specific mutation by a selected restriction enzyme that will not cleave the wildtype DNA. In a study using a mixture of wildtype and mutation containing DNA amplified by regular PCR or COLD-PCR, COLD-PCR preceding RFLP analysis was shown to improve the mutation detection by 10-20 fold.[2]
Sanger sequencing
Sanger sequencing recently was used to evaluate the enrichment of mutant DNA from a mixture of 1:20 mutant:wildtype DNA. The variant DNA containing a mutation was obtained from a breast cancer cell line known to contain p53 mutations.[1] Comparison of Sanger sequencing chromatograms indicated that the mutant allele was enriched 13 fold when COLD-PCR was used compared to traditional PCR alone.[1] This was determined by the size of the peaks on the chromatogram at the variant allele location.
As well, COLD-PCR was used to detect p53 mutations from lung-adenocarcinoma samples. The study was able to detect 8 low level (under 20% abundance) mutations that would likely have been missed using conventional methods that don't enrich for variant sequence DNA.
Pyrosequencing
Similar to its use in direct Sanger sequencing, with pyrosequencing COLD-PCR was shown to be capable of detecting mutations that had a prevalence 0.5–1% from the samples used.[3] COLD-PCR was used to detect p53 and KRAS mutations by pyrosequencing, and was shown to outperform conventional PCR in both cases.
MALDI-TOF
The same research group that developed COLD-PCR and used it to compare the sensitivity of regular PCR for genotyping with direct Sanger sequencing, RFLP, and pyrosequencing, also ran a similar study using MALDI-TOF as a downstream application for detecting mutations. Their results indicated that COLD-PCR could enrich mutation sequences from a mixture of DNA by 10–100 fold and that mutations with an initial prevalence of 0.1–0.5% would be detectable.[4] Compared to the 5–10% low-level detection rate expected with traditional PCR.
QPCR
COLD-PCR run on a quantitative PCR machine, using TaqMan probes specific for a mutation, was shown to increase the measured difference between mutant and wildtype samples.[4]
Advantages
- Single-step method capable of enriching both known and unknown minority alleles irrespective of mutation type and position
- Does not require extra costly reagents or specialized machinery
- Better than conventional PCR for the detection of mutations in a mixed sample
- Does not significantly increase experiment run time compared to conventional PCR
Disadvantages
- Optimal Tc must be measured and determined for each amplicon, adding an extra step to conventional PCR-based procedures
- Requirement for precise denaturation temperature control during PCR to within ± 0.3 °C (0.54 °F)
- A suitable critical temperature may not be available that differentiates between mutant and wildtype DNA sequences
- Restricted to analyzing sequences smaller than approximately 200bp
- Vulnerable to polymerase-introduced errors
- Variable overall mutation enrichment dependent on DNA position and nucleotide substitution
- No guarantee that all low-level mutations will be preferentially enriched
History
COLD-PCR was originally described by Li et al. in a Nature Medicine paper published in 2008 from Mike Makrigiorgos's lab group at the Dana Farber Cancer Institute of Harvard Medical School.[2] As summarized above, the technology has been used in a number of proof-of-principle experiments and medical research diagnostic experiments.
Recently, the COLD-PCR technology has been licensed by Transgenomic, Inc. The licensing terms include the exclusive rights to commercialize the technology combined with Sanger sequencing. The plans are to develop commercial applications that will allow for rapid high-sensitivity detection of low-level somatic and mitochondrial DNA mutations.[5]
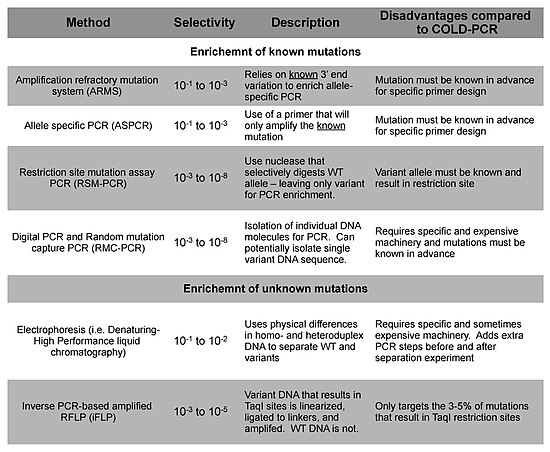
Alternatives
Other technologies are available for the detection of minority DNA mutations, and these methods can be segregated into their ability to enrich for and detect either known or unknown mutations.[6]
See also
References
- ^ a b c Li J, Milbury CA, Li C, Makrigiorgos GM (November 2009). "Two-round COLD-PCR-based Sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma". Human Mutation. 30 (11): 1583–90. doi:10.1002/humu.21112. PMC 2784016. PMID 19760750.
- ^ a b Li J, Wang L, Mamon H, Kulke MH, Berbeco R, Makrigiorgos GM (May 2008). "Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing". Nature Medicine. 14 (5): 579–84. doi:10.1038/nm1708. PMID 18408729. S2CID 205385306.
- ^ Zuo Z, Chen SS, Chandra PK, et al. (August 2009). "Application of COLD-PCR for improved detection of KRAS mutations in clinical samples". Modern Pathology. 22 (8): 1023–31. doi:10.1038/modpathol.2009.59. PMID 19430420.
- ^ a b Li J, Makrigiorgos GM (April 2009). "COLD-PCR: a new platform for highly improved mutation detection in cancer and genetic testing". Biochemical Society Transactions. 37 (2): 427–32. doi:10.1042/BST0370427. PMID 19290875.
- ^ Laboratorytalk editorial team (Oct 2009). "Transgenomics licenses COLD-PCR technology". Archived from the original on 2011-07-18. Retrieved 2010-03-04.
- ^ Milbury CA, Li J, Makrigiorgos GM (April 2009). "PCR-Based Methods for the Enrichment of Minority Alleles and Mutations". Clinical Chemistry. 55 (4): 632–40. doi:10.1373/clinchem.2008.113035. PMC 2811432. PMID 19201784.
