Aerolysin
| Aerolysin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
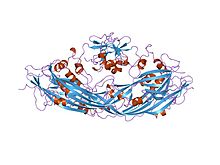 proaerolysin | |||||||||
| Identifiers | |||||||||
| Symbol | Aerolysin | ||||||||
| Pfam | PF01117 | ||||||||
| Pfam clan | CL0345 | ||||||||
| InterPro | IPR005830 | ||||||||
| PROSITE | PDOC00247 | ||||||||
| SCOP2 | 1pre / SCOPe / SUPFAM | ||||||||
| TCDB | 1.C.4 | ||||||||
| OPM superfamily | 35 | ||||||||
| OPM protein | 5jzt | ||||||||
| |||||||||
In molecular biology, aerolysin is a cytolytic pore-forming toxin exported by Aeromonas hydrophila, a Gram-negative bacterium associated with diarrhoeal diseases and deep wound infections.[1][2] It is also produced by the caterpillar of the moth Megalopyge opercularis, sometimes called the Tree Asp. The mature toxin binds to eukaryotic cells and aggregates to form holes (approximately 3 nm in diameter) leading to the destruction of the membrane permeability barrier and osmotic lysis. The structure of proaerolysin has been determined to 2.8A resolution and shows the protoxin to adopt a novel fold.[2] Images of an aerolysin oligomer derived from electron microscopy have helped to construct a model of the protein in its heptameric conformation, and to outline a mechanism by which this assembly might insert into lipid bilayers to form ion channels.[3]
References
- ^ Howard SP, Garland WJ, Green MJ, Buckley JT (June 1987). "Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila". J. Bacteriol. 169 (6): 2869–71. doi:10.1128/jb.169.6.2869-2871.1987. PMC 212202. PMID 3584074.
- ^ a b Parker MW, Buckley JT, Postma JP, Tucker AD, Leonard K, Pattus F, Tsernoglou D (January 1994). "Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states". Nature. 367 (6460): 292–5. doi:10.1038/367292a0. PMID 7510043. S2CID 4371932.
- ^ Degiacomi MT, Iacovache I, Pernot L, Chami M, Kudryashev M, Stahlberg H, van der Goot FG, Dal Peraro M (August 2013). "Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism. The gene for aerolysin have been shown to undergo Horizontal gene transfer from prokaryotes to eukaryotes" (PDF). Nature Chemical Biology. 9 (6460): 623–629. doi:10.1038/nchembio.1312. PMID 23912165.
