Tabtoxin
| |
| Names | |
|---|---|
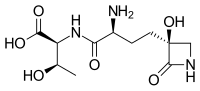
| Systematic IUPAC name (2S,3R)-2-{(2S)-2-Amino-4-[(3S)-3-hydroxy-2-oxoazetidin-3-yl]butanamido}-3-hydroxybutanoic acid | |
| Other names N-[(2S)-2-Amino-4-[(3S)-3-hydroxy-2-oxo-3-azetidinyl]-1-oxobutyl]-L-threonine; (S)-γ-(3-Hydroxy-2-oxo-3-azetidinyl)-L-α-aminobutyryl-L-threonine | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C11H19N3O6 | |
| Molar mass | 289.288 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
Tabtoxin, also known as wildfire toxin, is a simple monobactam phytotoxin produced by Pseudomonas syringae. It is the precursor to the antibiotic tabtoxinine β-lactam (TBL).[1] It is produced by:
- Pseudomonas syringae pv. tabaci, the causal agent of the wildfire of tobacco.
- P. syringae pv. coronafaciens
- P. syringae pv. garcae
- P. syringae BR2, causes a disease of bean (Phaseolus vulgaris) similar to tobacco wildfire. This organism is closely related to P. syringae pv. tabaci but cannot be classified in the pathovar tabaci because it is not pathogenic on tobacco.
Tabtoxin is a dipeptide precursor to the biologically active form of TBL, differing by having an extra threonine attached by a peptide bond to the C terminus. Tabtoxin is required by BR2(R) for both chlorosis and lesion formation on bean. All mutations that affected tabtoxin production, whether spontaneous deletion or transposon induced, also affected lesion formation, and in all cases, restoration of tabtoxin production also restored pathogenic symptoms. Other factors may be required for BR2 to be pathogenic on bean, but apparently these are in addition to tabtoxin production.[2][3] TBL functions as a toxin by inhibition of glutamine synthetase.
Self-resistance
Tabtoxin resistance protein (TTR) is an enzyme that catalyzes the acetylation of TBL, rendering tabtoxin-producing pathogens tolerant to their own phytotoxins. The structure of an inactivated mutant of TTR is solved with its natural cofactor acetyl-CoA to 1.55 Å resolution. The binary complex forms a characteristic “V” shape for substrate binding and contains the four motifs conserved in the GCN5-related N-acetyltransferase (GNAT) superfamily, which also includes the histone acetyltransferases (HATs).[4] There are reports that TTR possesses HAT activity and suggest an evolutionary relationship between TTR and other GNAT members.[2]
The production of tabtoxin itself is also part of the self-resistance strategy. These pathogens produce TBL before tabtoxin is produced; to detoxify, the enzyme TblF links TBL to threonine (Thr), producing a non-toxic product.[5]
Biosynthesis
The biosynthetic precursors of tabtoxin were identified by the incorporation of 13C-labeled compounds. L-threonine and L-aspartate make up the side chain, while pyruvic acid and the methyl group of L-methionine make up β-lactam moiety. A biosynthetic model for the formation of TβL resembles that of lysine, where the first dedicated step is the DapA-catalyzed condensation of aspartic acid semialdehyde with pyruvate to form L-2,3-dihydropicolinate (DHDPA). Tabtoxin biosynthesis branches off from the lysine biosynthetic pathway before the formation of diaminopimelate (DAP).[a] The 31-kb biosynthetic cluster consists of:[6]
- TabA (P31851), an enzyme related to lysA (diaminopimelate decarboxylase)
- TabB (P31852), an enzyme related to dapD (THDPA succinyl-CoA succinyltransferase, THDPA-ST)
- TblA (P31850), an enzyme with no close paralogs (identified as a member of SAMe-dependent methyltransferase superfamily by InterPro)
This pathway produces TBL; the enzyme TblF finalize the synthesis by linking TBL to Thr to form tabtoxin.[b][6]
Factors affecting production
The effects of carbon, nitrogen sources and amino acids on growth and tabtoxin production by pv. tabaci, were examined by varying the components of a defined basal medium, which contained the following nutrients per liter: sucrose (10 g), KNO3 (5 g), MgSO4.7H2O (0.2 g), CaCl2.2H2O (0.11 g), FeSO4.7H2O (20 mg), NaH2PO4.2H2O (0.9 g) and H2PO4.3H2O[clarification needed] (1 g). Both growth and quantity of tabtoxin synthesized were significantly affected by carbon source, nitrogen source and amino acid supplements. Sorbitol, xylose and sucrose proved to be the best carbon sources for tabtoxin production. Specific toxin production was very low using glucose as a single carbohydrate source, although bacterial growth was well supported by glucose. Amount and type of nitrogen sources (NH4Cl or KNO3) affected the growth of pv. tabaci and quantities of tabtoxin produced. Nitrate is the best of these two forms of nitrogen for production of tabtoxin.
Some progress has been made on elucidating factors that regulate tabtoxin biosynthesis in P. syringae.[6]
Bioactivation
Zinc was shown to be required for the aminopeptidase activity, which hydrolyzes tabtoxin to release TβL.[6]
References
- ^ As additional evidence for this pathway, DapB (Dihydrodipicolinate reductase), a step between DHDPA and DAP, is required to produce both lysine and tabtoxin.
- ^ TblF is able to distinguish between TBL and the spontaneously-formed isomer TDL (tabtoxinine-δ-lactam). It also produces no "reversed" peptides.
- ^ Kinscherf TG, Coleman RH, Barta TM, Willis DK (July 1991). "Cloning and expression of the tabtoxin biosynthetic region from Pseudomonas syringae". J. Bacteriol. 173 (13): 4124–32. doi:10.1128/jb.173.13.4124-4132.1991. PMC 208062. PMID 1648077.
- ^ a b Arai, Toshinobu; Arimura, Yasuhiro; Ishikura, Shun; Kino, Kuniki (15 August 2013). "l-Amino Acid Ligase from Pseudomonas syringae Producing Tabtoxin Can Be Used for Enzymatic Synthesis of Various Functional Peptides". Appl. Environ. Microbiol. 79 (16): 5023–5029. Bibcode:2013ApEnM..79.5023A. doi:10.1128/AEM.01003-13. PMC 3754701. PMID 23770908.
- ^ Kinscherf, T. G.; Coleman, R. H.; Barta, T. M.; Willis, D. K. (1 July 1991). "Cloning and expression of the tabtoxin biosynthetic region from Pseudomonas syringae". Journal of Bacteriology. 173 (13): 4124–4132. doi:10.1128/jb.173.13.4124-4132.1991. PMC 208062. PMID 1648077.
- ^ Rao, Yi Ding, Shentao Li, Xiaofeng Li, Fei Sun, Jinyuan Liu, Nanming Zhao and Zihe (31 May 2003). "Site-Directed Mutagenesis and Preliminary X-Ray Crystallographic Studies of the Tabtoxin Resistance Protein". Protein and Peptide Letters. 10 (3): 255–63. doi:10.2174/0929866033478924. PMID 12871145.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - ^ Wencewicz, TA; Walsh, CT (2 October 2012). "Pseudomonas syringae self-protection from tabtoxinine-β-lactam by ligase TblF and acetylase Ttr". Biochemistry. 51 (39): 7712–25. doi:10.1021/bi3011384. PMID 22994681.
- ^ a b c d Bender, Carol L.; Alarcón-Chaidez, Francisco; Gross, Dennis C. (1 June 1999). "Pseudomonas syringae Phytotoxins: Mode of Action, Regulation, and Biosynthesis by Peptide and Polyketide Synthetases". Microbiol. Mol. Biol. Rev. 63 (2): 266–292. doi:10.1128/MMBR.63.2.266-292.1999. PMC 98966. PMID 10357851.
- http://aem.asm.org/content/79/16/5023.long
- He, Hongzhen; Ding, Yi; Bartlam, Mark; Sun, Fei; Le, Yi; Qin, Xincheng; Tang, Hong; Zhang, Rongguang; Joachimiak, Andrzej; Liu, Jinyuan; Zhao, Nanming; Rao, Zihe (2003). "Crystal Structure of Tabtoxin Resistance Protein Complexed with Acetyl Coenzyme A Reveals the Mechanism for β-Lactam Acetylation". Journal of Molecular Biology. 325 (5): 1019–1030. doi:10.1016/S0022-2836(02)01284-6. PMID 12527305.
