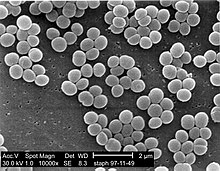
Staphylococcus
Staphylococcus is a genus of Gram-positive bacteria in the family Staphylococcaceae from the order Bacillales. Under the microscope, they appear spherical (cocci), and form in grape-like clusters. Staphylococcus species are facultative anaerobic organisms (capable of growth both aerobically and anaerobically).
The name was coined in 1880 by Scottish surgeon and bacteriologist Alexander Ogston (1844–1929), following the pattern established five years earlier with the naming of Streptococcus.[1] It combines the prefix "staphylo-" (from Ancient Greek: σταφυλή, romanized: staphylē, lit. 'bunch of grapes'[2]), and suffixed by the New Latin: coccus, lit. 'spherical bacterium' (from Ancient Greek: κόκκος, romanized: kókkos, lit. 'grain, seed, berry'[3]).
Staphylococcus was one of the leading infections in hospitals and many strains of this bacterium have become antibiotic resistant. Despite strong attempts to get rid of them, staphylococcus bacteria stay present in hospitals, where they can infect people who are most at risk of infection.[4]
Staphylococcus includes at least 44 species. Of these, nine have two subspecies, one has three subspecies, and one has four subspecies.[5] Many species cannot cause disease and reside normally on the skin and mucous membranes of humans and other animals. Staphylococcus species have been found to be nectar-inhabiting microbes.[6] They are also a small component of the soil microbiome.[7]
Taxonomy
The taxonomy is based on 16s rRNA sequences,[8] and most of the staphylococcal species fall into 11 clusters:
- S. aureus group – S. argenteus, S. aureus, S. schweitzeri, S. simiae
- S. auricularis group – S. auricularis
- S. carnosus group – S. carnosus, S. condimenti, S. debuckii, S. massiliensis, S. piscifermentans, S. simulans
- S. epidermidis group – S. capitis, S. caprae, S. epidermidis, S. saccharolyticus
- S. haemolyticus group – S. borealis, S. devriesei, S. haemolyticus, S. hominis
- S. hyicus-intermedius group – S. agnetis, S. chromogenes, S. cornubiensis, S. felis, S. delphini, S. hyicus, S. intermedius, S. lutrae, S. microti, S. muscae, S. pseudintermedius, S. rostri, S. schleiferi
- S. lugdunensis group – S. lugdunensis
- S. saprophyticus group – S. arlettae, S. caeli, S. cohnii, S. equorum, S. gallinarum, S. kloosii, S. leei, S. nepalensis, S. saprophyticus, S. succinus, S. xylosus
- S. sciuri group – S. fleurettii, S. lentus, S. sciuri, S. stepanovicii, S. vitulinus
- S. simulans group – S. simulans
- S. warneri group – S. pasteuri, S. warneri
A twelfth group – that of S. caseolyticus – has now been removed to a new genus, Macrococcus, the species of which are currently the closest known relatives of Staphylococcus.[9]
Two species were described in 2015 – Staphylococcus argenteus and Staphylococcus schweitzeri – both of which were previously considered variants of S. aureus.[10]
A new coagulase negative species – Staphylococcus edaphicus – has been isolated from Antarctica.[11] This species is probably a member of the S. saprophyticus group.
Groups
Based on an analysis of orthologous gene content three groups (A, B and C) have been proposed.[12]
Group A includes S. aureus, S. borealis, S. capitis, S. epidermidis, S. haemolyticus, S. hominis, S. lugdunensis, S. pettenkoferi, S. simiae and S. warneri.
Group B includes S. arlettae, S. cohnii, S. equorum, S. saprophyticus and S. xylosus.
Group C includes S. delphini, S. intermedius and S. pseudintermedius.
Notes
The S. saprophyticus and S. sciuri groups are generally novobiocin-resistant, as is S. hominis subsp. novobiosepticus.
Members of the S. sciuri group are oxidase-positive due to their possession of the enzyme cytochrome c oxidase. This group is the only clade within the staphylococci to possess this gene.
The S. sciuri group appears to be the closest relations to the genus Macrococcus.
S. pulvereri has been shown to be a junior synonym of S. vitulinus.[13]
Within these clades, the S. haemolyticus and S. simulans groups appear to be related, as do the S. aureus and S. epidermidis groups.[14]
S. lugdunensis appears to be related to the S. haemolyticus group.
S. petrasii may be related to S. haemolyticus, but this needs to be confirmed.
The taxonomic position of S. lyticans,S. petrasii, and S. pseudolugdunensis has yet to be clarified. The published descriptions of these species do not appear to have been validly published.
Biochemical identification
Assignment of a strain to the genus Staphylococcus requires it to be a Gram-positive coccus[15] that forms clusters, has an appropriate cell wall structure (including peptidoglycan type and teichoic acid presence) and G + C content of DNA in a range of 30–40 mol%.
Staphylococcus species can be differentiated from other aerobic and facultative anaerobic, Gram-positive cocci by several simple tests.[15] Staphylococcus species are facultative anaerobes (capable of growth both aerobically and anaerobically).[15] All species grow in the presence of bile salts.
All species of Staphylococcus aureus were once thought to be coagulase-positive, but this has since been disproven.[16][17][18]
Growth can also occur in a 6.5% NaCl solution.[15] On Baird-Parker medium, Staphylococcus species grow fermentatively, except for S. saprophyticus, which grows oxidatively. Staphylococcus species are resistant to bacitracin (0.04 U disc: resistance = < 10 mm zone of inhibition) and susceptible to furazolidone (100 μg disc: resistance = < 15 mm zone of inhibition). Further biochemical testing is needed to identify to the species level.
Coagulase production
One of the most important phenotypical features used in the classification of staphylococci is their ability to produce coagulase, an enzyme that causes blood clot formation.
Seven species are currently recognised as being coagulase-positive: S. aureus, S. delphini, S. hyicus, S. intermedius, S. lutrae, S. pseudintermedius, and S. schleiferi subsp. coagulans. These species belong to two separate groups – the S. aureus (S. aureus alone) group and the S. hyicus-intermedius group (the remaining five).
An eighth species has also been described – Staphylococcus leei – from patients with gastritis.[19]
S. aureus is coagulase-positive, meaning it produces coagulase. However, while the majority of S. aureus strains are coagulase-positive, some may be atypical in that they do not produce coagulase. S. aureus is catalase-positive (meaning that it can produce the enzyme catalase) and able to convert hydrogen peroxide (H2O2) to water and oxygen, which makes the catalase test useful to distinguish staphylococci from enterococci and streptococci.
S. pseudintermedius inhabits and sometimes infects the skin of domestic dogs and cats. This organism, too, can carry the genetic material that imparts multiple bacterial resistance. It is rarely implicated in infections in humans, as a zoonosis.
S. epidermidis, a coagulase-negative species, is a commensal of the skin, but can cause severe infections in immunosuppressed patients and those with central venous catheters. S. saprophyticus, another coagulase-negative species that is part of the normal vaginal flora, is predominantly implicated in genitourinary tract infections in sexually active young women. In recent years, several other Staphylococcus species have been implicated in human infections, notably S. lugdunensis, S. schleiferi, and S. caprae.
Common abbreviations for coagulase-negative staphylococci are CoNS, CNS, or CNST.[20] The American Society for Microbiology abbreviates coagulase-negative staphylococci as "CoNS".
Genomics and molecular biology
The first S. aureus genomes to be sequenced were those of N315 and Mu50, in 2001. Many more complete S. aureus genomes have been submitted to the public databases, making it one of the most extensively sequenced bacteria. The use of genomic data is now widespread and provides a valuable resource for researchers working with S. aureus. Whole genome technologies, such as sequencing projects and microarrays, have shown an enormous variety of S. aureus strains. Each contains different combinations of surface proteins and different toxins. Relating this information to pathogenic behaviour is one of the major areas of staphylococcal research. The development of molecular typing methods has enabled the tracking of different strains of S. aureus. This may lead to better control of outbreak strains. A greater understanding of how the staphylococci evolve, especially due to the acquisition of mobile genetic elements encoding resistance and virulence genes is helping to identify new outbreak strains and may even prevent their emergence.[21]
The widespread incidence of antibiotic resistance across various strains of S. aureus, or across different species of Staphylococcus has been attributed to horizontal gene transfer of genes encoding antibiotic/metal resistance and virulence. A recent study demonstrated the extent of horizontal gene transfer among Staphylococcus to be much greater than previously expected, and encompasses genes with functions beyond antibiotic resistance and virulence, and beyond genes residing within the mobile genetic elements.[22]
Various strains of Staphylococcus are available from biological research centres, such as the National Collection of Type Cultures.
Host range
Members of the genus Staphylococcus frequently colonize the skin and upper respiratory tracts of mammals and birds and also in marine sponge.[15] Marine sponge associated Staphylococcus species are highly salt tolerant.[15] Some species specificity has been observed in host range, such that the Staphylococcus species observed on some animals appear more rarely on more distantly related host species.[23] Some of the observed host specificity includes:
- S. arlattae – chickens, goats, marine sponge[15]
- S. aureus – humans,[24] cattle
- S. auricularis – deer, dogs, humans
- S. borealis – humans, cattle
- S. capitis – humans
- S. caprae – goats, humans
- S. cohnii – chickens, humans
- S. delphini – dolphins
- S. devriesei – cattle
- S. epidermidis – humans, marine sponge[15]
- S. equorum – horses
- S. felis – cats
- S. fleurettii – goats
- S. gallinarum – chickens, goats, pheasants
- S. haemolyticus – humans, Cercocebus, Erythrocebus, Lemur, Macca, Microcebus, Pan
- S. hyicus – pigs
- S. leei – humans
- S. lentus – goats, rabbits, sheep
- S. lugdunensis – humans, goats
- S. lutrae – otters
- S. microti – voles (Microtus arvalis)
- S. nepalensis – goats
- S. pasteuri – humans, goats
- S. pettenkoferi – humans
- S. pseudintermedius – dogs
- S. rostri – pigs
- S. schleiferi – humans
- S. sciuri – humans, dogs, goats
- S. simiae – South American squirrel monkeys (Saimiri sciureus)
- S. simulans – humans
- S. warneri – humans, Cercopithecoidea, Pongidae
- S. xylosus – humans
Populations at risk for Staphylococcus aureus infection
It is said that anyone can develop a staph infection, although certain groups of people are at greater risk, including people with chronic conditions such as diabetes, cancer, vascular disease, eczema, lung disease, and people who inject drugs. In healthcare facilities, the risk of more serious staph infection is higher because many patients have weakened immune systems or have undergone procedures. In healthcare, the risk of more serious staph infection is higher for patients in intensive care units (ICUs), patients who have undergone certain types of surgeries and patients with medical devices inserted in their bodies.[25]
Staphylococcus aureus has emerged as a leading agent of sepsis. It facilitates factors such as tissue adhesion, immune evasion, and host cell injury. In the bloodstream, these factors cause inflammation, impair immune cell function, alter coagulation, and compromise vascular integrity. When left untreated, S. aureus triggers pathophysiologic disturbances that are further amplified by the host inflammatory response, culminating in the severe clinical manifestations of sepsis and septic shock.[26]
Clinical
Staphylococcus can cause a wide variety of diseases in humans and animals through either toxin production or penetration. Staphylococcal toxins are a common cause of food poisoning, for they can be produced by bacteria growing in improperly stored food items. The most common sialadenitis is caused by staphylococci, as bacterial infections.[27] Staphylococci break down leucine into isovaleric acid, the main odor of foot odor.[28]
See also
References
- ^ "staphylococcus | Origin and meaning of staphylococcus by Online Etymology Dictionary". www.etymonline.com. Retrieved 2018-07-25.
- ^ stafulh/ in Liddell, Henry George; Scott, Robert (1940) A Greek–English Lexicon, revised and augmented throughout by Jones, Sir Henry Stuart, with the assistance of McKenzie, Roderick. Oxford: Clarendon Press. In the Perseus Digital Library, Tufts University.
- ^ ko)kkos in Liddell and Scott
- ^ "Staph infections". mayoclinic.org. Retrieved 2022-11-27.
- ^ Harris LG, Foster SJ, Richards RG (December 2002). "An introduction to Staphylococcus aureus, and techniques for identifying and quantifying S. aureus adhesins in relation to adhesion to biomaterials: review". European Cells & Materials. 4: 39–60. doi:10.22203/ecm.v004a04. PMID 14562246.
- ^ Jacquemyn H, Lenaerts M, Brys R, Willems K, Honnay O, Lievens B (2013). "Among-population variation in microbial community structure in the floral nectar of the bee-pollinated forest herb Pulmonaria officinalis L." PLOS ONE. 8 (3): e56917. Bibcode:2013PLoSO...856917J. doi:10.1371/journal.pone.0056917. PMC 3594240. PMID 23536759.
- ^ Madigan M, Martinko J, eds. (2005). Brock Biomlogy of Microorganisms (11th ed.). Prentice Hall. ISBN 978-0-13-144329-7.[page needed]
- ^ Takahashi T, Satoh I, Kikuchi N (April 1999). "Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis". International Journal of Systematic Bacteriology. 49 (2): 725–8. doi:10.1099/00207713-49-2-725. PMID 10319495.
- ^ Kloos WE, Ballard DN, George CG, Webster JA, Hubner RJ, Ludwig W, Schleifer KH, Fiedler F, Schubert K (July 1998). "Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., and Macrococcus bovicus sp. no. and Macrococcus carouselicus sp. nov". International Journal of Systematic Bacteriology. 48 (3): 859–77. doi:10.1099/00207713-48-3-859. PMID 9734040.
- ^ Tong SY, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM (January 2015). "Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov". International Journal of Systematic and Evolutionary Microbiology. 65 (Pt 1): 15–22. doi:10.1099/ijs.0.062752-0. PMC 4298100. PMID 25269845.
- ^ Pantůček R, Sedláček I, Indráková A, Vrbovská V, Mašlaňová I, Kovařovic V, Švec P, Králová S, Krištofová L, Kekláková J, Petráš P, Doškař J (October 2017). "mecCgene and genomic islands with suspected role in adaptation to extreme environment". Applied and Environmental Microbiology. 84 (2): e01746–17. doi:10.1128/AEM.01746-17. PMC 5752872. PMID 29079617.
- ^ Coates-Brown R, Moran JC, Pongchaikul P, Darby AC and MJ Horsburgh MJ (2018) "Comparative genomics of Staphylococcus reveals determinants of speciation and diversification of antimicrobial defense". Front Microbiol
- ^ Svec P, Vancanneyt M, Sedlácek I, Engelbeen K, Stetina V, Swings J, Petrás P (November 2004). "Reclassification of Staphylococcus pulvereri Zakrzewska-Czerwinska et al. 1995 as a later synonym of Staphylococcus vitulinus Webster et al. 1994". International Journal of Systematic and Evolutionary Microbiology. 54 (Pt 6): 2213–5. doi:10.1099/ijs.0.63080-0. PMID 15545460.
- ^ Ghebremedhin B, Layer F, König W, König B (March 2008). "Genetic classification and distinguishing of Staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences". Journal of Clinical Microbiology. 46 (3): 1019–25. doi:10.1128/JCM.02058-07. PMC 2268370. PMID 18174295.
- ^ a b c d e f g h Paul, Sulav Indra; Rahman, Md. Mahbubur; Salam, Mohammad Abdus; Khan, Md. Arifur Rahman; Islam, Md. Tofazzal (2021-12-15). "Identification of marine sponge-associated bacteria of the Saint Martin's island of the Bay of Bengal emphasizing on the prevention of motile Aeromonas septicemia in Labeo rohita". Aquaculture. 545: 737156. doi:10.1016/j.aquaculture.2021.737156. ISSN 0044-8486.
- ^ Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 978-0-8385-8529-0.[page needed]
- ^ PreTest, Surgery, 12th ed., p. 88
- ^ Matthews KR, Roberson J, Gillespie BE, Luther DA, Oliver SP (1997). "Identification and Differentiation of Coagulase-Negative Staphylococcus aureus by Polymerase Chain Reaction". Journal of Food Protection. 60 (6): 686–8. doi:10.4315/0362-028X-60.6.686. PMID 31195568.
- ^ Jin M, Rosario W, Watler E, Calhoun DH (March 2004). "Development of a large-scale HPLC-based purification for the urease from Staphylococcus leei and determination of subunit structure". Protein Expression and Purification. 34 (1): 111–7. doi:10.1016/j.pep.2003.10.012. PMID 14766306.
- ^ Becker K, Heilmann C, Peters G (October 2014). "Coagulase-negative staphylococci". Clinical Microbiology Reviews. 27 (4): 870–926. doi:10.1128/CMR.00109-13. PMC 4187637. PMID 25278577.
- ^ Lindsay J, ed. (2008). Staphylococcus: Molecular Genetics. Caister Academic Press. ISBN 978-1-904455-29-5.[page needed]
- ^ Chan CX, Beiko RG, Ragan MA (August 2011). "Lateral transfer of genes and gene fragments in Staphylococcus extends beyond mobile elements". Journal of Bacteriology. 193 (15): 3964–77. doi:10.1128/JB.01524-10. PMC 3147504. PMID 21622749.
- ^ Kloos WE (1980). "Natural populations of the genus Staphylococcus". Annual Review of Microbiology. 34: 559–92. doi:10.1146/annurev.mi.34.100180.003015. PMID 7002032.
- ^ "Symptoms and Treatments of Staph Infection (Cellulitis)". Just-Health.net. December 14, 2013.
- ^ "Staphylococcus aureus in Healthcare Settings | HAI". CDC. 2020-12-10. Retrieved 2022-04-23.
- ^ Powers, Michael E.; Wardenburg, Juliane Bubeck (13 February 2014). "Igniting the Fire: Staphylococcus aureus Virulence Factors in the Pathogenesis of Sepsis". PLOS Pathogens. 10 (2): e1003871. doi:10.1371/journal.ppat.1003871. ISSN 1553-7374. PMC 3923759. PMID 24550724.
- ^ "Sialoadenitis: inflammation of the salivary glands". The Medical Consumer's Advocate. 2001-01-04. Retrieved 2011-01-04.
- ^ Stevens D, Cornmell R, Taylor D, Grimshaw SG, Riazanskaia S, Arnold DS, Fernstad SJ, Smith AM, Heaney LM, Reynolds JC, Thomas CL, Harker M. Spatial variations in the microbial community structure and diversity of the human foot is associated with the production of odorous volatiles. FEMS Microbiol Ecol. 2015 Jan;91(1):1-11. doi: 10.1093/femsec/fiu018. Epub 2014 Dec 8. PMID: 25764539.
External links
 Media related to Staphylococcus at Wikimedia Commons
Media related to Staphylococcus at Wikimedia Commons Data related to Staphylococcus at Wikispecies
Data related to Staphylococcus at Wikispecies- Staphylococcus genomes and related information at PATRIC, a Bioinformatics Resource Center funded by NIAID