Exon junction complex
An exon junction complex (EJC) is a protein complex which forms on a pre-messenger RNA strand at the junction of two exons which have been joined together during RNA splicing. The EJC has major influences on translation, surveillance, localization of the spliced mRNA, and m6A methylation. [1] [2] It is first deposited onto mRNA during splicing and is then transported into the cytoplasm. There it plays a major role in post-transcriptional regulation of mRNA. It is believed that exon junction complexes provide a position-specific memory of the splicing event. The EJC consists of a stable heterotetramer core, which serves as a binding platform for other factors necessary for the mRNA pathway.[2] The core of the EJC contains the protein eukaryotic initiation factor 4A-III (eIF4A-III; a DEAD-box RNA helicase) bound to an adenosine triphosphate (ATP) analog, as well as the additional proteins Magoh and Y14.[3] The binding of these proteins to nuclear speckled domains has been measured recently and it may be regulated by PI3K/AKT/mTOR signaling pathways.[4] In order for the binding of the complex to the mRNA to occur, the eIF4AIII factor is inhibited, stopping the hydrolysis of ATP.[3] This recognizes EJC as an ATP dependent complex. EJC also interacts with a large number of additional proteins; most notably SR proteins.[5] These interactions are suggested to be important for mRNA compaction.[5] The role of EJC in mRNA export is controversial.
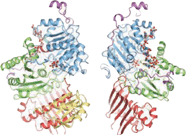
Protein components
The EJC is made up of several key protein components: RNPS1, Y14, SRm160, Aly/REF and Magoh, among others.[6][7][8] RNPS1 can function as a coactivator of splicing, but along with Y14, it also takes part in the process of nonsense-mediated decay in eukaryotes.[9][10] SRm160 is another coactivator that has been proposed to enhance mRNA 3’ end processing.[11][12] The protein component Magoh is thought to facilitate the subcytoplasmic localization of mRNAs while Aly is engaged in nuclear mRNA export.[13][14][15] Aly is believed to be recruited to the exon junction complex by the protein UAP56.[16] UAP56 is recognized as an RNA helicase but acts as a splicing factor required for early spliceosome assembly.[17] Another factor involved in the EJC pathway is DEK. This component is known to take part in a variety of functions ranging from splicing to transcriptional regulation and chromatin structure.[18][19][20]
Structure
The crystallization of the exon junction complex has revealed the structural organization of its protein components. The core of the complex is elongated with an overall dimension of 99Å by 67Å by 54Å.[21] It is organized around the eIF4AIII factor. The factor itself consists of two different types of conformations around the mRNA: closed and open. In a closed state, the two domains of eIF4AIII form composite binding sites for the 5'-adenylyl-β,γ-imidodiphosphate (ADPNP) and mRNA.[21] In the open conformation, the two domains are rotated by 160 degrees relative to closed state.[18] The protein components Magoh and Y14 bind together to form a heterodimer located at the 5’ pole of the EJC.[22][23][24] Magoh binds to an eIF4AIII domain through interactions between residues from its two C-terminal helices and one end of a large β-sheet.[21] Conserved residues in the linker between the two eIF4AIII domains form salt bridges or hydrogen bonds with specific residues in Magoh.[21] Other bonding occurs between the second loop of the Magoh β–sheet and the two eIF4AIII domains and their linker.[21] There is only a single partial bond formed between Y14 and eIF4AIII. This consists of a salt bridge between the conserved residues Y14 Arg108 and eIF4AIII Asp401.[21] If mutations were to occur to both of these residues, association of Magoh-Y14 with EJC would be non-existent.[25]
Mechanism
During the second step of splicing in eukaryotic cells, the EJC is deposited approximately 20-24 nucleotides from the 5’ end upstream of the splice junction (where two exons are joined), when the lariat has formed and the exons are ligated together.[26][27] The binding of the EJC to the mRNA occurs in a sequence independent manner, to form the mature messenger ribonucleoprotein (mRNP).[28] The EJC remains stably bound to this mRNP as it is exported out of the nucleus and into the cytoplasm. Protein components are either bound to or released by the EJC as it is transported. In order for the translocation of mRNAs through the nuclear pore complex to occur, a heterodimer consisting of NXF1/TAP and NXT1/p15 must bind to the transcripts.[29] NXF1/TAP is a major receptor for the export of mRNAs to the cytoplasm. This is because it interacts with both RNA-binding adapter proteins and components of the nuclear pore complex.[30]
Recognition of a premature termination codon occurs during translation in the cytoplasm. The image shown below implies that this event is nuclear, contrary to the general view in this field. Readers should be aware that translation in the nucleus is a highly controversial subject that is not well-supported by data.[citation needed]

In nonsense mediated decay
Exon junction complexes play a major role in mRNA surveillance. More specifically, they are found in the nonsense mediated decay pathway (NMD), wherein mRNA transcripts with premature stop codons are degraded. In normal mRNA translation, the ribosome binds to the transcript and begins amino acid chain elongation. It continues on until it reaches the location of the exon junction complex, which it then displaces. Next, translation is complete when the ribosome reaches a termination codon. In NMD, the mRNA transcript contains a premature termination codon (PTC) due to a nonsense mutation. If this codon occurs prior to the EJC site, the EJC will remain bound, triggering mRNA decay.[31] The EJC and its position serve as a type of regulator, determining whether the transcript is defective or not.
EJCs are also known to take part in NMD in another way; the recruitment of the surveillance factors UPF1, UPF2 and UPF3.[32] These proteins are the most important components of the NMD mechanism. The EJC protein MAGOH, Y14 and eIF4AIII provide a binding for UPF3, which acts as a bridge between UPF2 and UPF1 forming a trimeric complex.[33] Within this complex, UPF2 and UPF3 act cooperatively to promote ATPase and RNA helicase of UPF1.[33] The EJC core stably anchors the UPF complex to the mRNA, and aids in regulation of essential UPF1 protein.[33] Ribosomes which are stalled on a PTC recruit UPF1 through interactions with the release factor eRF1 and eRF3.[33] Along with the protein SMG1, eRF1, eRF3 and UPF1 form the complex SURF. This complex forms a bridge between the ribosome and the downstream EJC which is associated with UPF3 and UPF2.[33] This interaction triggers the phosphorylation of UPF1 by SMG1, causing the dissociation of eRF1 and eRF3.[33] The complex produced consists of EJC, UPF3, UPF2, phosphorylated UPF1 and SMG1 and in turn triggers degradation of the mRNA.[33]
References
- ^ He, P Cody; Wei, Jiangbo; Dou, Xiaoyang; Harada, Bryan; Zhang, Zijie; Ge, Ruiqi; Liu, Chang; Zhang, Li-sheng; Yu, Xianbin; Wang, Shuai; Lyu, Ruitu; Zou, Zhongyu; Chen, Mengjie; He, Chuan (February 2023). "Exon architecture controls mRNA m6A suppression and gene expression". Science. 379 (6633): 677–682. doi:10.1126/science.abj9090. PMC 9990141. PMID 36705538.
- ^ a b Tange, TØ; Nott, A; Moore, MJ (June 2004). "The ever-increasing complexities of the exon junction complex". Current Opinion in Cell Biology. 16 (3): 279–84. doi:10.1016/j.ceb.2004.03.012. PMID 15145352.
- ^ a b Ballut L, Marchadier B, Baguet A, Tomasetto C, Séraphin B, Le Hir H (2005). "The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity". Nat. Struct. Mol. Biol. 12 (10): 861–9. doi:10.1038/nsmb990. PMID 16170325. S2CID 1359792.
- ^ Quaresma, Alexandre J.; Nickerson, Jeffrey A. (2013), "Regulation of mRNA export by the PI3 kinase/AKT signal transduction pathway", Mol Biol Cell, 24 (8): 1208–21, doi:10.1091/mbc.E12-06-0450, PMC 3623641, PMID 23427269
- ^ a b Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ (2012). "The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus". Cell. 151 (4): 750–64. doi:10.1016/j.cell.2012.10.007. PMC 3522173. PMID 23084401.
- ^ Kataoka, N.; Yong, J.; Kim, V. N.; Velazquez, F.; Perkinson, R. A.; Wang, F.; Dreyfuss, G. (2000). "Pre-mRNA Splicing Imprints mRNA in the Nucleus with a Novel RNA-Binding Protein that Persists in the Cytoplasm". Mol Cell. 6 (3): 673–682. doi:10.1016/s1097-2765(00)00065-4. PMID 11030346.
- ^ Le Hir H, Gatfield D, Izaurralde E, Moore MJ (2001). "The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay". EMBO J. 20 (17): 4987–97. doi:10.1093/emboj/20.17.4987. PMC 125616. PMID 11532962.
- ^ Le Hir, H.; Izaurralde, E.; Maquat, L. E.; Moore, M. J. (2000). "The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions". EMBO J. 19 (24): 6860–6869. doi:10.1093/emboj/19.24.6860. PMC 305905. PMID 11118221.
- ^ Lykke-Andersen, J.; Shu, M.-D.; Steitz, J. A. (2001). "Communication of the Position of Exon-Exon Junctions to the mRNA Surveillance Machinery by the Protein RNPS1". Science. 293 (5536): 1836–1839. Bibcode:2001Sci...293.1836L. doi:10.1126/science.1062786. PMID 11546874. S2CID 389385.
- ^ Lejeune, F.; Ishigaki, Y.; Li, X.; Maquat, L. E. (2002). "The exon junction complex is detected on CBP80-bound but not eIF4E-bound mRNA in mammalian cells: dynamics of mRNP remodeling". EMBO J. 21 (13): 3536–3545. doi:10.1093/emboj/cdf345. PMC 126094. PMID 12093754.
- ^ Mayeda, A.; Badolato, J.; Kobayashi, R.; Zhang, M. Q.; Gardiner, E. M.; Krainer, A. R. (1999). "Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing". EMBO J. 18 (16): 4560–4570. doi:10.1093/emboj/18.16.4560. PMC 1171530. PMID 10449421.
- ^ McCracken, S.; Lambermon, M.; Blencowe, B. J. (2002). "SRm160 Splicing Coactivator Promotes Transcript 3'-End Cleavage". Mol. Cell. Biol. 22 (1): 148–160. doi:10.1128/mcb.22.1.148-160.2002. PMC 134228. PMID 11739730.
- ^ Le Hir, H.; Gatfield, D.; Braun, I. C.; Forler, D.; Izaurralde, E. (2001). "The protein Mago provides a link between splicing and mRNA localization". EMBO Rep. 2 (12): 1119–1124. doi:10.1093/embo-reports/kve245. PMC 1084163. PMID 11743026.
- ^ Zhou, Z.; Luo, M.-J.; Straesser, K.; Katahira, J.; Hurt, E.; Reed, R. (2000). "The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans". Nature. 407 (6802): 401–405. Bibcode:2000Natur.407..401Z. doi:10.1038/35030160. PMID 11014198. S2CID 312262.
- ^ Rodrigues, J. P.; Rode, M.; Gatfield, D.; Blencowe, B. J.; Carmo-Fonseca, M.; Izaurralde, E. (2001). "REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus". Proc. Natl. Acad. Sci. USA. 98 (3): 1030–1035. Bibcode:2001PNAS...98.1030R. doi:10.1073/pnas.98.3.1030. PMC 14703. PMID 11158589.
- ^ Cullen, B. R. (2003). "Nuclear RNA export". J. Cell Sci. 116 (4): 587–597. doi:10.1242/jcs.00268. PMID 12538759.
- ^ Gatfield, D.; Izaurralde, E. (2002). "REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export". J. Cell Biol. 159 (4): 579–588. doi:10.1083/jcb.200207128. PMC 2173090. PMID 12438415.
- ^ a b Alexiadis V, Waldmann T, Andersen J, Mann M, Knippers R, Gruss C (2000). "The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner". Genes Dev. 14 (11): 1308–12. doi:10.1101/gad.14.11.1308. PMC 316669. PMID 10837023.
- ^ McGarvey, T.; Rosonina, E.; McCracken, S.; Li, Q.; Arnaout, R.; Mientjes, E.; Nickerson, J.A.; Awrey, D.; Greenblatt, J.; Grosveld, G.; Blencowe, BJ (2000). "The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes". J. Cell Biol. 150 (2): 309–320. doi:10.1083/jcb.150.2.309. PMC 2180225. PMID 10908574.
- ^ Faulkner, N.E.; Hilfinger, J.M.; Markovitz, D.M. (2001). "Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site". J. Biol. Chem. 276 (28): 25804–25812. doi:10.1074/jbc.m006454200. PMID 11320078.
- ^ a b c d e f Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Séraphin B, Le Hir H, Andersen GR (2006). "Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA". Science. 313 (5795): 1968–72. Bibcode:2006Sci...313.1968A. doi:10.1126/science.1131981. PMID 16931718. S2CID 26409491.
- ^ Lau, C. K.; Diem, M. D.; Dreyfuss, G.; Van Duyne, G. D. (2003). "Structure of the Y14-Magoh Core of the Exon Junction Complex". Curr. Biol. 13 (11): 933–941. Bibcode:2003CBio...13..933L. doi:10.1016/s0960-9822(03)00328-2. PMID 12781131.
- ^ Fribourg, S.; Gatfield, D.; Izaurralde, E. Conti (2003). "A novel mode of RBD-protein recognition in the Y14–Mago complex". Nat. Struct. Biol. 10 (6): 433–439. doi:10.1038/nsb926. PMID 12730685. S2CID 40116577.
- ^ Shi H, Xu RM (2003). "Crystal structure of the Drosophila Mago nashi-Y14 complex". Genes Dev. 17 (8): 971–6. doi:10.1101/gad.260403. PMC 196043. PMID 12704080.
- ^ Gehring, Niels H.; Kunz, Joachim B.; Neu-Yilik, Gabriele; Breit, Stephen; Viegas, Marcelo H.; Hentze, Matthias W.; Kulozik, Andreas E. (2005). "Exon-Junction Complex Components Specify Distinct Routes of Nonsense-Mediated mRNA Decay with Differential Cofactor Requirements". Molecular Cell. 20 (1): 65–75. doi:10.1016/j.molcel.2005.08.012. ISSN 1097-2765. PMID 16209946.
- ^ Reichert, V.L.; Le Hir, H.; Jurica, M.S.; Moore, M.J. (2002). "5' exon interactions within the spliceosome establish a framework for exon junction complex structure and assembly". Genes Dev. 16 (21): 2778–2791. doi:10.1101/gad.1030602. PMC 187475. PMID 12414731.
- ^ Shibuya, T.; Tange, T.O.; Sonenberg, N.; Moore, M.J. (2004). "eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense mediated decay". Nat. Struct. Mol. Biol. 11 (4): 346–351. doi:10.1038/nsmb750. PMID 15034551. S2CID 30171314.
- ^ Le Hir, H.; Izaurralde, E.; Maquat, L.E.; Moore, M.J. (2000). "The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions". EMBO J. 19 (24): 6860–6869. doi:10.1093/emboj/19.24.6860. PMC 305905. PMID 11118221.
- ^ Reed, R.; Hurt, E. (2002). "A conserved mRNA export machinery coupled to pre-mRNA splicing". Cell. 108 (4): 523–531. doi:10.1016/s0092-8674(02)00627-x. PMID 11909523.
- ^ Izaurralde, E (2002). "A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur". J. Cell Biol. 81 (11): 577–584. doi:10.1078/0171-9335-00273. PMID 12498157.
- ^ Chang YF, Imam JS, Wilkinson MF (2007). "The nonsense-mediated decay RNA surveillance pathway". Annu Rev Biochem. 76: 51–74. doi:10.1146/annurev.biochem.76.050106.093909. PMID 17352659.
- ^ Conti, E.; Izaurralde, E. (2005). "Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species". Curr. Opin. Cell Biol. 17 (3): 316–325. doi:10.1016/j.ceb.2005.04.005. PMID 15901503.
- ^ a b c d e f g Chamieh, Hala; Ballut, Lionel; Bonneau, Fabien; Le Hir, Hervé (2007). "NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity". Nature Structural & Molecular Biology. 15 (1): 85–93. doi:10.1038/nsmb1330. ISSN 1545-9993. PMID 18066079. S2CID 6268216.
