CrysTBox
 | |
| Developer(s) | Miloslav Klinger |
|---|---|
| Initial release | 9 December 2014 |
| Written in | MATLAB |
| Operating system | Microsoft Windows |
| Available in | English |
| Type | Scientific |
| License | Free for non-commercial use |
| Website | crystbox.fzu.cz |
CrysTBox (Crystallographic Tool Box) is a suite of computer tools designed to accelerate material research based on transmission electron microscope images via highly accurate automated analysis and interactive visualization. Relying on artificial intelligence and computer vision, CrysTBox makes routine crystallographic analyses simpler, faster and more accurate compared to human evaluators. The high level of automation together with sub-pixel precision[1] and interactive visualization makes the quantitative crystallographic analysis accessible even for non-crystallographers allowing for an interdisciplinary research. Simultaneously, experienced material scientists can take advantage of advanced functionalities for comprehensive analyses.
CrysTBox is being developed in the Laboratory of electron microscopy at the Institute of Physics of the Czech Academy of Sciences. For academic purposes, it is available for free. As of 2022, the suite has been deployed at research and educational facilities in more than 90 countries[2] supporting research of ETH Zurich,[3] Lawrence Berkeley National Laboratory,[4] Max Planck Institutes,[5] Chinese Academy of Sciences,[6] Fraunhofer Institutes[7] or Oxford University.[8]
Suite
As a scientific tool, CrysTBox suite is freely available for academic purposes, it supports file formats widely used in the community and offers interconnection with other scientific software.
Availability
CrysTBox is freely available on demand for non-commercial use by non-commercial subjects. The only safe way to download CrysTBox installers is via a request form on the official website.[note 1] Commercial use is not allowed due to the license of MATLAB used for CrysTBox compilation.[2]
Notable research and users
Besides education, CrysTBox is mainly used in research with fields of application spanning from nuclear research[9] to archaeology and paleontology.[10] Among others, the suite was employed in development of additive manufacturing (including 3D printed biodegradable alloys,[7] metallic glass[11] or high-entropy alloys[12]), resistant coatings,[13] laser shock peening,[14] water cleaning technologies[15] or characterization of 50 million years old flint.[10]
Institutions whose research was supported by CrysTBox include educational facilities such as ETH Zurich,[3] University of California,[16] Uppsala University,[13] Oxford University,[8] University of Waterloo,[15] Indian Institute of Technology,[17] Nanyang Technological University[12] or University of Tokyo[18] as well as research institutes like Max Planck Institutes,[12] Chinese Academy of Sciences,[6] Fraunhofer Institutes[7] or US national laboratories (NL) such as Oak Ridge NL,[9] Lawrence Berkeley NL,[4] Idaho NL[9] and Lawrence Livermoore NL.[14]
Limitations and disadvantages
CrysTBox is compiled to a stand-alone installers using MATLAB Compiler. Therefore, 1-2 GB of MATLAB libraries are installed together with the toolbox.
The diffraction simulation used in cellViewer is based on kinematic diffraction theory. This allows for a real-time response to user interaction, but it does not cover advanced diffraction features like double diffraction covered by dynamical diffraction theory, even though some phenomena caused by multiple electron-matter interactions are visualized by CrysTBox - for instance Kikuchi lines.
The analytical tools provide correction for scale calibration imperfections, but does not provide adjustment for image distortions such as elliptical distortion. If high-accuracy measurement is needed or if the distortion exceeds standard levels, appropriate tools[19] should be applied prior to the analysis.
Crystallographic visualization tools
In order to visualize functional relations and provide better understanding of experimental data, the graphical interface emphasize user interactivity and functional interconnection. There are two visualization tools in the suite: one depicting single material while another being focused on intergrowths of two different materials.
cellViewer - single crystal visualization
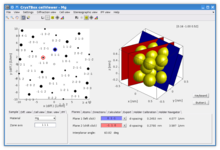
CellViewer allows to visualize the sample material in four modes widely used in material research:
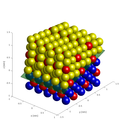
- 3D model of atomic structure (direct space),
- simulated diffraction pattern (reciprocal space),
- stereographic projection (projection of 3D space of crystallographic planes and directions to 2D),
- inverse pole figure (defined part of stereographic projection).
Graphical user interface provides user with two interactive views side by side. These views can display arbitrary combination of the four aforementioned visualization modes allowing to perceive their mutual relations. For instance, rotation of the atomic structure in direct space leads (if set so) to an instant update of the simulated diffraction pattern. If any diffraction spot is selected, corresponding crystallographic planes are shown in the unit cell etc. Such interconnections are implemented for each pair of the four available visualization modes. The electronic visualization allows to simplify understanding of widely used, yet less intuitive representations such as the inverse pole figure. For instance by drawing the coloured triangle of the inverse pole figure into the stereographic projection or to the more intuitive 3D atomic structure.[1]
- Examples of various usage of visualization modes with magnesium as sample material
- 3D atomic structure with selected planes
- Diffraction pattern with selected reflections (planes)
- 3D atomic structure with inverse pole figure
- Stereographic projection with inverse pole figure
- Inverse pole figure
ifaceViewer - intergrowth visualization

The ifaceViewer allows for visualization of two misoriented materials and their interface such as crystal twins or grain boundaries. The user interface provides three views: two smaller views, each depicting one unit cell of selected material and orientation, and a larger view depicting an appropriate interface of the two structures. The interface can be visualized in four modes:
- 3D model of both unit cells,
- wire-frame model of both unit cells,
- cross section of the interface,
- bulk representation (up to several hundred atoms).
All three views in the user interface are functionally interconnected. If the content of one view is rotated by the user, the other views follow. If a crystallographic plane or direction is selected in one view, it is shown in other views and corresponding crystallographic indices are stated. The tool also allows to highlight coincident site lattice or calculate the list of planes and directions which are parallel or nearly parallel in the two misoriented materials.[1]
- Interface of two aluminum lattices in Σ5 misorientation shown in four available visualization modes of CrysTBox IfaceViewer
- 3D model of both unit cells
- Wire model of both unit cells
- Interface cross section
- Bulk representation
Automated analysis of TEM images
CrysTBox offers tools for automated processing of diffraction patterns and high-resolution transmission electron microscope images. Since the tools employ algorithms of artificial intelligence and computer vision, they are designed to require minimal operator effort providing higher accuracy compared to manual evaluation. Four analytical tools can be used to index diffraction patterns, measure lattice constants (distances and angles), sample thickness etc. Despite the high level of automation, the user is able to control the whole process and perform individual steps manually if needed.
diffractGUI - HRTEM and diffraction processing

DiffractGUI allows for an automated analysis of diffraction patterns and high-resolution images of single crystal or limited number of crystallites. It is able to determine crystal orientation, index individual diffraction spots and measure interplanar angles and distances in picometric precision.[20] The input image may depict:
- selected area diffraction pattern,
- high-resolution image,
- nanodiffraction pattern or
- convergent beam electron diffraction.
The input image is processed in the following steps:
- Preprocessing with accordance to the settings and image nature (resolution and noise reduction, Fourier transform for direct space images etc.).
- Detection of diffraction reflections at various scales[21] (difference of Gaussians typically used for spot detection, Hough transform[22][23] for CBED disk detection[24]).
- The strongest detections are selected across the scale space.
- A regular lattice is fit to the set of the strongest detections using RANSAC[25] algorithm.
- Lengths and angles of the lattice basis vectors are measured.
- Crystal lattice orientation is determined and diffraction reflections are identified using theoretical parameters of the sample material.[26][27]
Compared to human evaluation, diffractGUI considers tens or even hundreds of diffraction spots at once and, therefore, can localize the pattern with sub-pixel precision.[20]
- Individual steps of automated procedure illustrated on diffraction pattern of titanium
- Input image
- 30 strongest reflections detected
- Regular lattice
- Lattice basis and parameters measured
- Reflections identified for zone axis [0 -1 1]
ringGUI - ring diffraction analysis
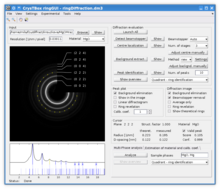
RingGUI allows for an automated processing of ring diffraction images of polycrystalline or powder samples. It can be used to identify the diffraction rings, quantify the interplanar distances and thus characterize or identify the sample material. With known material, it can assist in microscope calibration. The input image is processed as follows:
- beam-stopper detection,
- localization of the ring center,
- quantification of the diffraction profile and estimation of its background intensity,
- identification of the rings[27] in the image (peaks in the profile).
The results can be further processed and visualized in two interactive, functionally interconnected graphical elements:
- Interactive diffraction image – allows the user to improve readability of the diffraction image by removing the beam-stopper, subtracting the background, revealing faint or spotty rings or by crystallographic identification of the depicted rings.
- Diffraction profile – circular average of the image intensities depicts the peaks corresponding to the rings and their match with theoretical values known for given sample material.
Both, the diffraction image as well as diffraction profile can be used to select diffraction rings with a mouse click. The corresponding ring is then highlighted in both graphical representations and details are listed.[1]
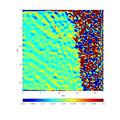
- Examples of patrtial and final results presented via interactive diffraction image of MgO
- Input image
- Beam-stopper detected
- Ring centre localized
- Background removed
- Beam-stopper removed
- Background and beam-stopper removed with faint rings revealed
- Rings identified
- Comparison of experimental and theoretical rings
twoBeamGUI - sample thickness estimation
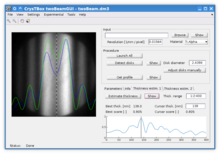
Sample thickness can be estimated using twoBeamGUI from a convergent beam electron diffraction pattern (CBED) in two beam approximation.[26][28] The procedure is based on an automated extraction of the intensity profile across the diffracted disk in the following steps:
- diffraction disk radius is determined using multi-scale Hough transform,[24]
- the transmitted and diffracted disks are localized and the reflection is indexed,[26][27]
- the disks are horizontally aligned, cropped out and profiles are measured across the disks,
- the profile across the diffracted disk is matched with a series of profiles automatically simulated[26] for given material, reflection and specified thickness range.
Once the procedure is completed, the measured profile and the most similar simulated profile are displayed with the diffracted disk on the background. This allows the user to verify correctness of the automated estimate and easily check the similarity of other intensity profiles within the specified thickness range.[1]
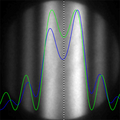
- Individual steps of automated thickness estimation of titanium sample
- Input image
- Disks detected
- Disks aligned with profiles measured
- Comparison of measured (blue) and simulated (green) profile for thickness of 139 nm
gpaGUI - geometric phase analysis

The tool called gpaGUI provides an interactive interface for geometric phase analysis. It allows to generate 2D maps of various crystallographic quantities using high-resolution images.[29][30]
Since the geometric phase analysis is performed in frequency domain, the high-resolution image needs to be transformed into frequential representation using Fourier transform. Mathematically, the frequential image is a complex matrix with the size equal to the original image. Crystallographically, it can be seen as an artificial diffraction pattern of the original image depicting intensity peaks corresponding to the crystallographic planes present in the original image. After performing desired calculations, the frequential representation can be transformed back to the original spatial domain using inverse Fourier transform.
Various crystallographic analyses can be performed using the frequential image. If it is filtered so that only the information from a region close to a particular diffraction spot is used (the rest is set to zero), a filtered direct image obtained by inverse Fourier transform then depicts only the planes corresponding to the selected diffraction spot. Moreover, due to its complex nature, the frequential image can be used to calculate amplitude and phase. Together with a vector of one crystallographic plane depicted in the image, they can be used to generate a 2D map interplanar distance of given plane.[29] If two vectors of non-parallel planes are known, the method can be used to generate maps of strain and displacement.[30]
Graphical user interface of gpaGUI is vertically divided into two halves, each of which contains:
- Diffractogram preview allowing to select one diffraction spot corresponding to a crystallographic plane.
- Visualization of a selected quantity (input image, filtered image or one of the maps mentioned above) allowing to select point of interest or region of interest for further analysis.
- Results of detailed analysis of point or region of interest. The point analysis allows the user to select any pixel of the visualized map to see exact values of the particular pixel and its closest neighbourhood. If analysis of broader area is needed, a polygonal region can be outlined in the map allowing to enumerate its statistical details: mean, standard deviation, median, minimum, maximum and total area of the polygon.
Since each half of the interface allows to specify one crystallographic plane, gpaGUI allows to calculate all the aforementioned crystallographic quantities including those which require two vectors. Precision and repeatability of the whole analysis relies on accuracy of the diffraction peak localization. To overcome inaccuracy of manual peak localization (with a mouse click), gpaGUI provides a possibility to process the input image with diffractGUI in order to accurately localize and index the peaks.[1]
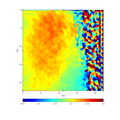
- Examples of 2D maps generated using gpaGUI from high resolution image of magnesium.
- Input image
- Filtered image of plane (0 1 0)
- Map of (0 1 0) d-spacing
- Map of XX component of strain tensor
- Map of X component of displacement vector
See also
- Transmission electron microscope
- Selected area diffraction
- Convergent beam electron diffraction
- High-resolution transmission electron microscopy
- Geometric phase analysis
- Electron crystallography
- Crystal structure
- Computer vision
- Artificial intelligence
- Fourier transform
- Difference of gaussians
- Hough transform
- RANSAC
- Czech Academy of Sciences
Notes
- ^ CrysTBox is not distributed by any website aggregating various software installers or uninstallers. Installers downloaded from these sources can contain malware. Similarly, no additional software is required to uninstall CrysTBox. Any software pretending to do so has no connection to CrysTBox developer.
References
- ^ a b c d e f Klinger, Miloslav (2017-07-07). "More features, more tools, more CrysTBox". Journal of Applied Crystallography. 50 (4). International Union of Crystallography (IUCr): 1226–1234. doi:10.1107/s1600576717006793. ISSN 1600-5767.
- ^ a b "CrysTBox - Crystallographic Toolbox". Fyzikální ústav Akademie věd České republiky. 2021-11-16.
- ^ a b Cihova, M.; Schäublin, R.; Hauser, L.B.; Gerstl, S.S.A.; Simson, C.; Uggowitzer, P.J.; Löffler, J.F. (2018). "Rational design of a lean magnesium-based alloy with high age-hardening response". Acta Materialia. 158. Elsevier BV: 214–229. Bibcode:2018AcMat.158..214C. doi:10.1016/j.actamat.2018.07.054. ISSN 1359-6454. S2CID 139448116.
- ^ a b Schoeppler, Vanessa; Stier, Deborah; Best, Richard J.; Song, Chengyu; Turner, John; Savitzky, Benjamin H.; Ophus, Colin; Marcus, Matthew A.; Zhao, Shiteng; Bustillo, Karen; Zlotnikov, Igor (2021). "Crystallization by Amorphous Particle Attachment: On the Evolution of Texture". Advanced Materials. 33 (37). Wiley: 2101358. Bibcode:2021AdM....3301358S. doi:10.1002/adma.202101358. ISSN 0935-9648. PMC 11468020. PMID 34337782. S2CID 236777450.
- ^ Sandoval-Diaz, Luis; Plodinec, Milivoj; Ivanov, Danail; Poitel, Stéphane; Hammud, Adnan; Nerl, Hannah C.; Schlögl, Robert; Lunkenbein, Thomas (2020). "Visualizing the importance of oxide-metal phase transitions in the production of synthesis gas over Ni catalysts". Journal of Energy Chemistry. 50. Elsevier BV: 178–186. Bibcode:2020JEnCh..50..178S. doi:10.1016/j.jechem.2020.03.013. hdl:21.11116/0000-0006-4374-C. ISSN 2095-4956. S2CID 216467746.
- ^ a b Ma, Jiqiang; Fan, Chenglong; Chen, Wenyuan; Tan, Hui; Zhu, Shengyu; Li, Qinglin; Yang, Jun (2022). "Core-shell structure in situ reinforced aluminum matrix composites: Microstructure, mechanical and tribological properties". Journal of Alloys and Compounds. 901. Elsevier BV: 163613. doi:10.1016/j.jallcom.2022.163613. ISSN 0925-8388. S2CID 245719646.
- ^ a b c Bär, Florian; Berger, Leopold; Jauer, Lucas; Kurtuldu, Güven; Schäublin, Robin; Schleifenbaum, Johannes H.; Löffler, Jörg F. (2019). "Laser additive manufacturing of biodegradable magnesium alloy WE43: A detailed microstructure analysis". Acta Biomaterialia. 98. Elsevier BV: 36–49. doi:10.1016/j.actbio.2019.05.056. ISSN 1742-7061. PMID 31132536. S2CID 167220852.
- ^ a b Hong, Zuliang; Morrison, Alasdair P. C.; Zhang, Hongtao; Roberts, Steve G.; Grant, Patrick S. (2017-12-04). "Development of a Novel Melt Spinning-Based Processing Route for Oxide Dispersion-Strengthened Steels". Metallurgical and Materials Transactions A. 49 (2). Springer Science and Business Media LLC: 604–612. doi:10.1007/s11661-017-4398-x. ISSN 1073-5623. S2CID 58942346.
- ^ a b c Cappia, F.; Winston, A.; Miller, B.; Kombaiah, B.; Teng, F.; Murray, D.; Frazer, D.; Harp, J.M. (2021). "Electron microscopy characterization of the fuel-cladding interaction in medium burnup annular fast reactor MOX". Journal of Nuclear Materials. 551. Elsevier BV: 152922. Bibcode:2021JNuM..55152922C. doi:10.1016/j.jnucmat.2021.152922. ISSN 0022-3115. OSTI 1782045. S2CID 233835199.
- ^ a b Natalio, Filipe; Corrales, Tomas P.; Pierantoni, Maria; Rosenhek-Goldian, Irit; Cernescu, Adrian; Raguin, Emeline; Maria, Raquel; Cohen, Sidney R. (2021). "Characterization of Eocene flint". Chemical Geology. 582. Elsevier BV: 120427. Bibcode:2021ChGeo.58220427N. doi:10.1016/j.chemgeo.2021.120427. ISSN 0009-2541.
- ^ Marattukalam, Jithin James; Pacheco, Victor; Karlsson, Dennis; Riekehr, Lars; Lindwall, Johan; Forsberg, Fredrik; Jansson, Ulf; Sahlberg, Martin; Hjörvarsson, Björgvin (2020). "Development of process parameters for selective laser melting of a Zr-based bulk metallic glass". Additive Manufacturing. 33. Elsevier BV: 101124. doi:10.1016/j.addma.2020.101124. ISSN 2214-8604. S2CID 213366256.
- ^ a b c Sun, Zhongji; Tan, Xipeng; Wang, Chengcheng; Descoins, Marion; Mangelinck, Dominique; Tor, Shu Beng; Jägle, Eric A.; Zaefferer, Stefan; Raabe, Dierk (2021). "Reducing hot tearing by grain boundary segregation engineering in additive manufacturing: example of an AlxCoCrFeNi high-entropy alloy" (PDF). Acta Materialia. 204. Elsevier BV: 116505. doi:10.1016/j.actamat.2020.116505. ISSN 1359-6454. S2CID 228880026.
- ^ a b Malinovskis, Paulius; Fritze, Stefan; Riekehr, Lars; von Fieandt, Linus; Cedervall, Johan; Rehnlund, David; Nyholm, Leif; Lewin, Erik; Jansson, Ulf (2018). "Synthesis and characterization of multicomponent (CrNbTaTiW)C films for increased hardness and corrosion resistance". Materials & Design. 149. Elsevier BV: 51–62. doi:10.1016/j.matdes.2018.03.068. ISSN 0264-1275. S2CID 103441835.
- ^ a b Zhao, Shiteng; Kad, Bimal; Remington, Bruce A.; LaSalvia, Jerry C.; Wehrenberg, Christopher E.; Behler, Kristopher D.; Meyers, Marc A. (2016-10-12). "Directional amorphization of boron carbide subjected to laser shock compression". Proceedings of the National Academy of Sciences. 113 (43): 12088–12093. Bibcode:2016PNAS..11312088Z. doi:10.1073/pnas.1604613113. ISSN 0027-8424. PMC 5087058. PMID 27733513.
- ^ a b Gora, Stephanie; Liang, Robert; Zhou, Y. Norman; Andrews, Susan (2018). "Settleable engineered titanium dioxide nanomaterials for the removal of natural organic matter from drinking water". Chemical Engineering Journal. 334. Elsevier BV: 638–649. Bibcode:2018ChEnJ.334..638G. doi:10.1016/j.cej.2017.10.058. hdl:10012/12609. ISSN 1385-8947.
- ^ Schuler, Jennifer D.; Rupert, Timothy J. (2017). "Materials selection rules for amorphous complexion formation in binary metallic alloys". Acta Materialia. 140. Elsevier BV: 196–205. arXiv:1708.02971. Bibcode:2017AcMat.140..196S. doi:10.1016/j.actamat.2017.08.042. ISSN 1359-6454. S2CID 20661118.
- ^ Sunkari, U.; Reddy, S. R.; Rathod, B. D. S.; Kumar, S. S. Satheesh; Saha, R.; Chatterjee, S.; Bhattacharjee, P. P. (2020-04-08). "Heterogeneous precipitation mediated heterogeneous nanostructure enhances strength-ductility synergy in severely cryo-rolled and annealed CoCrFeNi2.1Nb0.2 high entropy alloy". Scientific Reports. 10 (1). Springer Science and Business Media LLC: 6056. doi:10.1038/s41598-020-63038-z. ISSN 2045-2322. PMC 7142141. PMID 32269272.
- ^ Nakayama, Kei; Ishikawa, Ryo; Kobayashi, Shunsuke; Shibata, Naoya; Ikuhara, Yuichi (2020-09-08). "Dislocation and oxygen-release driven delithiation in Li2MnO3". Nature Communications. 11 (1). Springer Science and Business Media LLC: 4452. doi:10.1038/s41467-020-18285-z. ISSN 2041-1723. PMC 7479600. PMID 32901015.
- ^ Dave Mitchell (2022-02-05). "Ellipse fitting analysis".
- ^ a b Klinger, Miloslav; Polívka, Leoš; Jäger, Aleš; Tyunina, Marina (2016-04-12). "Quantitative analysis of structural inhomogeneity in nanomaterials using transmission electron microscopy". Journal of Applied Crystallography. 49 (3). International Union of Crystallography (IUCr): 762–770. doi:10.1107/s1600576716003800. ISSN 1600-5767.
- ^ Lowe, David G. (2004). "Distinctive Image Features from Scale-Invariant Keypoints". International Journal of Computer Vision. 60 (2). Springer Science and Business Media LLC: 91–110. doi:10.1023/b:visi.0000029664.99615.94. ISSN 0920-5691. S2CID 221242327.
- ^ Atherton, T.J.; Kerbyson, D.J. (1999). "Size invariant circle detection". Image and Vision Computing. 17 (11). Elsevier BV: 795–803. doi:10.1016/s0262-8856(98)00160-7. ISSN 0262-8856.
- ^ Yuen, HK; Princen, J; Illingworth, J; Kittler, J (1990). "Comparative study of Hough Transform methods for circle finding". Image and Vision Computing. 8 (1). Elsevier BV: 71–77. doi:10.1016/0262-8856(90)90059-e. ISSN 0262-8856.
- ^ a b Klinger, M.; Němec, M.; Polívka, L.; Gärtnerová, V.; Jäger, A. (2015). "Automated CBED processing: Sample thickness estimation based on analysis of zone-axis CBED pattern". Ultramicroscopy. 150. Elsevier BV: 88–95. doi:10.1016/j.ultramic.2014.12.006. ISSN 0304-3991. PMID 25544679.
- ^ Martin A. Fischler & Robert C. Bolles (June 1981). "Random Sample Consensus: A Paradigm for Model Fitting with Applications to Image Analysis and Automated Cartography" (PDF). Comm. ACM. 24 (6): 381–395. doi:10.1145/358669.358692. S2CID 972888. Archived (PDF) from the original on December 10, 2014.
- ^ a b c d Morniroli, Jean Paul (2004). Large-Angle Convergent-Beam Electron Diffraction Applications to Crystal Defects. Taylor & Francis. doi:10.1201/9781420034073. ISBN 978-2-901483-05-2.
- ^ a b c De Graef, Marc (2003-03-27). Introduction to Conventional Transmission Electron Microscopy. Cambridge University Press. doi:10.1017/cbo9780511615092. ISBN 978-0-521-62006-2.
- ^ Allen, Samuel M. (1981). "Foil thickness measurements from convergent-beam diffraction patterns". Philosophical Magazine A. 43 (2). Informa UK Limited: 325–335. Bibcode:1981PMagA..43..325A. doi:10.1080/01418618108239412. ISSN 0141-8610.
- ^ a b Hÿtch, M.J. (1997). "Geometric phase analysis of high resolution electron microscope images". Scanning Microscopy. 11: 53–66.
- ^ a b Hÿtch, M.J.; Snoeck, E.; Kilaas, R. (1998). "Quantitative measurement of displacement and strain fields from HREM micrographs". Ultramicroscopy. 74 (3). Elsevier BV: 131–146. doi:10.1016/s0304-3991(98)00035-7. ISSN 0304-3991.










![Reflections identified for zone axis [0 -1 1]](https://upload.wikimedia.org/wikipedia/commons/thumb/b/b2/DiffractGUI_reflections.png/120px-DiffractGUI_reflections.png)