Framework region
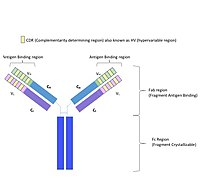
In molecular biology, a framework region is a subdivision of the variable region (Fab) of the antibody. The variable region is composed of seven amino acid regions, four of which are framework regions and three of which are hypervariable regions.[1] The framework region makes up about 85% of the variable region.[2] Located on the tips of the Y-shaped molecule, the framework regions are responsible for acting as a scaffold for the complementarity determining regions (CDR), also referred to as hypervariable regions, of the Fab. These CDRs are in direct contact with the antigen and are involved in binding antigen, while the framework regions support the binding of the CDR to the antigen[3] and aid in maintaining the overall structure of the four variable domains on the antibody.[4] To increase its stability, the framework region has less variability in its amino acid sequences compared to the CDR.[2]
Function
The antibody has a three-dimensional structure with beta pleated sheet and alpha helices.[5] The antibody folds so the variable regions form three loops with the framework regions folded into one another and the CDR regions on the tips of each of these loops in direct contact with the antigen.[5][6][7] Individual residues in the framework region can be divided into two categories, depending on whether they are in direct contact with the antigen. Framework residues that come in contact with the antigen are a part of the antibody's binding site, and are located either close in sequence to the CDRs or in close proximity to the CDR when in the folded three dimensional structure.[4] Framework residues that do not come in contact with the antigen affect the binding indirectly by aiding in structural support for the CDR. This enables the CDR to take on the correct orientation and position so it is exposed on the surface of the chain ready to bind to an antigen.[2]
The framework regions are highly conserved regions of the variable portion of the antibody. The evolutionary reason for the conservation of these regions is to support proper folding of the antibody allowing the CDR regions to be stabilized. Folding in FR leads to antibody structure flexibility or rigidity of the binding region of the antibody.[8][9]
Mutations
Mutations in the framework regions of antibodies occur in cells by somatic hypermutation and during affinity maturation of the antibody. In vitro, mutations of FR may occur by natural cause or by exposure to mutagens.[8][9] Recent studies of framework mutations imply that the framework region flexibility or rigidity could alter the specificity of the antibody to its intended epitope. While the framework region doesn't directly interact with antigen, its structure determines whether the CDRs can interact with antigen. If the CDR regions have high affinity for the epitope of antigen, it has been found to be more effective to have a more rigid framework region. When CDR does not have high affinity for antigen, mutations in the FR that create a more flexible structure may allow for higher affinity maturation.[8]
Natural mutations in the variable region are typically due to activation-induced cytidine deaminase (AID). AID leads to deamination of cytosine to uracil in DNA and results in somatic hypermutation. This somatic hypermutation allows for immunoglobulin class switching but also results in affinity maturation of the antibody. The CDR are the areas of the variable regions in contact with antigen and thus we see the most mutation in these regions. Although, the framework regions of the antibody are also mutated. Studies have shown that when the CDR is blocked from mutation and only the FR is mutated, certain mutations can lead to increased expression and thermostability of the antibody as a whole.[9] Antibody humanization is an example of beneficial genetic engineering in medicine today.[10] Humanized antibody refers to the creation of non-human antibody in vivo and in response to antigen, then the isolation and humanization of the framework and constant regions. It has been discovered that while these antibodies remain relatively intact upon transition, these modifications can also lead to decreased binding affinity in the humanized framework regions and result in improper folding in humans. This observation is believed to be due to the framework region's role in antibody structure.[10]
See also
References
- ^ "Antibody Structure". www.biology.arizona.edu. Retrieved 2018-01-16.
- ^ a b c Elgert, Klaus (1998). Immunology: Understanding the Immune System. John Wiley & Sons, Inc. p. 63.
- ^ Ill, C. R.; Gonzales, J. N.; Houtz, E. K.; Ludwig, J. R.; Melcher, E. D.; Hale, J. E.; Pourmand, R.; Keivens, V. M.; Myers, L. (1997-08-01). "Design and construction of a hybrid immunoglobulin domain with properties of both heavy and light chain variable regions". Protein Engineering. 10 (8): 949–957. doi:10.1093/protein/10.8.949. ISSN 0269-2139. PMID 9415445.
- ^ a b Sela-Culang, Inbal; Kunik, Vered; Ofran, Yanay (2013-10-08). "The Structural Basis of Antibody-Antigen Recognition". Frontiers in Immunology. 4: 302. doi:10.3389/fimmu.2013.00302. ISSN 1664-3224. PMC 3792396. PMID 24115948.
- ^ a b Zhu, Kai; Day, Tyler; Warshaviak, Dora; Murrett, Colleen; Friesner, Richard; Pearlman, David (2014-08-01). "Antibody structure determination using a combination of homology modeling, energy-based refinement, and loop prediction". Proteins: Structure, Function, and Bioinformatics. 82 (8): 1646–1655. doi:10.1002/prot.24551. ISSN 1097-0134. PMC 5282925. PMID 24619874.
- ^ Stanfield, Robyn L.; Wilson, Ian A. (2015-01-01). "Antibody Structure". In Crowe; Boraschi; Rappuoli (eds.). Antibodies for Infectious Diseases. pp. 49–62. doi:10.1128/9781555817411. ISBN 9781555817350.
- ^ Charles A Janeway, Jr; Travers, Paul; Walport, Mark; Shlomchik, Mark J. (2001). "The structure of a typical antibody molecule".
{{cite journal}}: Cite journal requires|journal=(help) - ^ a b c Ovchinnikov, Victor; Louveau, Joy E; Barton, John P; Karplus, Martin; Chakraborty, Arup K (2018-02-14). "Role of framework mutations and antibody flexibility in the evolution of broadly neutralizing antibodies". eLife. 7. doi:10.7554/elife.33038. ISSN 2050-084X. PMC 5828663. PMID 29442996.
- ^ a b c Lombana, T. Noelle; Dillon, Michael; III, Jack Bevers; Spiess, Christoph (2015-12-03). "Optimizing antibody expression by using the naturally occurring framework diversity in a live bacterial antibody display system". Scientific Reports. 5 (1): 17488. Bibcode:2015NatSR...517488L. doi:10.1038/srep17488. ISSN 2045-2322. PMC 4668361. PMID 26631978.
- ^ a b Caldas, Cristina; Coelho, Verônica; Kalil, Jorge; Moro, Ana Maria; Maranhão, Andrea Q; Brı́gido, Marcelo M (2003). "Humanization of the anti-CD18 antibody 6.7: an unexpected effect of a framework residue in binding to antigen". Molecular Immunology. 39 (15): 941–952. doi:10.1016/s0161-5890(03)00022-1. PMID 12695120.
